For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NPSVATKSVNARLAEELAIAEAQVVAAVRLLDEGATVPFIARYRKEATGSLDDGQLRTLEERLRYLRELD ERRAAVLASIEEQGKLTDELRATLMAVDTKARVEDIYLPYKPKRRTKAQIAREAGLEPLADRLLSDPTLA PESAAAEFISDDVADAAAALEGARHILVERAAEDAELVGAIREKFWSDASLQTAPWSADVVTTAAAQKFR DYFEFSERLHTMPSHRVLAVLRGEKEKVLSLNFDCGEDSPYEAMVAATLDIDLTAKAAVTPWLAATVSFA WRIRLISSATIDARVRLRQRAEVDAVSVFAKNLKDLLLAAPAGARTTLGLDPGFRTGVKVAVVDGTGKVV DTCAIYPHQPQRQWDTAKATLAALIARHRVNLIAIGNGTASREPDALATELISDIRGAGATPPAKAVVSE AGASVYSASAYAAHELPELDVTLRGAVSIARRLQDPLAELVKIEPKSIGVGQYQHDVTPGILARSLDAVV EDAVNAVGVDLNTASVPLLARVSGVSESLAEAIVAYRDNTGPFHNRRALLDVPRLGPKAFEQCAGFLRIR DGEDPLDASGVHPEAYPVVRRILDRTGVALAEIIGDERSLRSLRPADFADDRFGVPTVTDILAELEKPGR DPRPAFITATIAAGVEKVADLKVGMVLEGVVTNVAAFGAFVDVGVHQDGLVHVAAMAERFVSDPHEVVRS GQVVRVKVVDVDVERQRVGLSLRLNDEPQRDRANRADRAPKPRGGADSKLRRDQRNPHHAKCAGRRDSSS SGSSMAQALRDAGFGK*
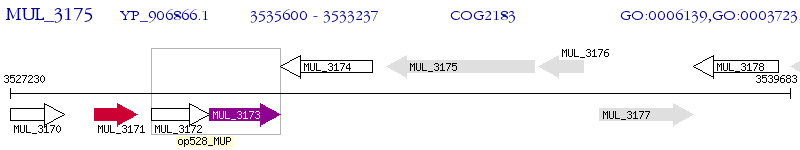
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3175 | tex | - | 100% (787) | transcriptional accessory protein Tex |
| M. ulcerans Agy99 | MUL_1612 | rpsA | 1e-13 | 41.67% (96) | 30S ribosomal protein S1 |
| M. ulcerans Agy99 | MUL_2150 | gpsI | 2e-05 | 30.93% (97) | polynucleotide phosphorylase/polyadenylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1656 | rpsA | 1e-13 | 45.12% (82) | 30S ribosomal protein S1 |
| M. gilvum PYR-GCK | Mflv_3444 | - | 0.0 | 80.10% (784) | RNA-binding S1 domain-containing protein |
| M. tuberculosis H37Rv | Rv1630 | rpsA | 1e-13 | 45.12% (82) | 30S ribosomal protein S1 |
| M. leprae Br4923 | MLBr_01382 | rpsA | 8e-14 | 45.12% (82) | 30S ribosomal protein S1 |
| M. abscessus ATCC 19977 | MAB_1549c | - | 0.0 | 78.45% (789) | RNA-binding protein |
| M. marinum M | MMAR_2585 | tex | 0.0 | 98.73% (787) | transcriptional accessory protein Tex |
| M. avium 104 | MAV_3152 | rpsA | 2e-13 | 45.12% (82) | 30S ribosomal protein S1 |
| M. smegmatis MC2 155 | MSMEG_2839 | - | 0.0 | 78.06% (793) | transcriptional accessory protein |
| M. thermoresistible (build 8) | TH_0210 | - | 0.0 | 78.26% (791) | transcriptional accessory protein |
| M. vanbaalenii PYR-1 | Mvan_3200 | - | 0.0 | 80.05% (782) | RNA-binding S1 domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3175|M.ulcerans_Agy99 -------MNPS---VATKSVNARLAEELAIAEAQVVAAVRLLDEGATVPF
MMAR_2585|M.marinum_M -------MNPS---VATKSVNARLAEELAIAEAQVVAAVRLLDEGATVPF
MAB_1549c|M.abscessus_ATCC_199 -------MTQS---VTVKSVNARLAAELAVTEAQVAAAVRLLDEGATVPF
MSMEG_2839|M.smegmatis_MC2_155 -------MPTSGPVPGVRSITARLAEELSVGEHQVAAAIHLLDEGSTVPF
TH_0210|M.thermoresistible__bu -------VTGT---VTLKSVNARLAEELEVGEAQVAAAVRLLDEGATVPF
Mflv_3444|M.gilvum_PYR-GCK MITSGAVKSGPAKSDAVKSVTARLAEELAVAEAQVAAAVRLLDEGATVPF
Mvan_3200|M.vanbaalenii_PYR-1 --MGGHSKS-SRVPRVIKSVNVRLAEELEVAETQVAAAVRLLDEGATVPF
Mb1656|M.bovis_AF2122/97 ----------------------------MPSPTVTSPQVAVNDIGSSEDF
Rv1630|M.tuberculosis_H37Rv ----------------------------MPSPTVTSPQVAVNDIGSSEDF
MAV_3152|M.avium_104 ----------------------------MPSPAVTSPQVAVNDIGSSEDF
MLBr_01382|M.leprae_Br4923 ----------------------------MSIPAVPSPQIAVNDVGSSEDF
. : : * *:: *
MUL_3175|M.ulcerans_Agy99 IARYRKEATGSLDDGQLRTLEERLRYLRELDERRAAVLASIEEQGKLTDE
MMAR_2585|M.marinum_M IARYRKEATGSLDDGQLRTLEERLRYLRELDERRAAVLASIEEQGKLTDE
MAB_1549c|M.abscessus_ATCC_199 IARYRKEVTGSLDDGQLRELEERLRYLRELDERRAAVLSSIEEQGKLTDE
MSMEG_2839|M.smegmatis_MC2_155 IARYRKEVTGSLDDGQLRTLEERLAYLRELDARREAVLASIEEQGKLTDE
TH_0210|M.thermoresistible__bu IARYRKEATGSLDDGQLRMLEERLRYLRELDERRAAVLSAIEEQGKLTDD
Mflv_3444|M.gilvum_PYR-GCK IARYRKEVTGSLDDGQLRALEERLGYLRELDDRRNAVLASINEQGKLTAE
Mvan_3200|M.vanbaalenii_PYR-1 IARYRKEATGSLDDGQLRVLEERLAYLRELDERRSAVLASIKEQGKLTDD
Mb1656|M.bovis_AF2122/97 LA----------------AIDKTIKYFNDGD-------------------
Rv1630|M.tuberculosis_H37Rv LA----------------AIDKTIKYFNDGD-------------------
MAV_3152|M.avium_104 LA----------------AIDKTIKYFNDGD-------------------
MLBr_01382|M.leprae_Br4923 LA----------------AIDKTIKYFNDGD-------------------
:* ::: : *:.: *
MUL_3175|M.ulcerans_Agy99 LRATLMAVDTKARVEDIYLPYKPKRRTKAQIAREAGLEPLADRLLSDPTL
MMAR_2585|M.marinum_M LRATLMAADTKARVEDIYLPYKPKRRTKAQIAREAGLEPLADRLLSDPTL
MAB_1549c|M.abscessus_ATCC_199 LKAALISADTKARVEDIYLPYKPKRRTKAQIAREAGLEPLADRLLADPNL
MSMEG_2839|M.smegmatis_MC2_155 LRAALVAADTKARVEDIYLPYKPKRRTKAQIAREAGLEPLADRLLADPNL
TH_0210|M.thermoresistible__bu LRAALLAADTKARVEDIYLPFKPKRRTKAQIAREAGLEPLADRLLADPTL
Mflv_3444|M.gilvum_PYR-GCK LTAALMAADTKARVEDIYLPYKPKRRTKAQIAREAGLQPLAERLLADPSV
Mvan_3200|M.vanbaalenii_PYR-1 LTAALMAADTKARVEDIYLPFKPKRRTKAQIAREAGLEPLADRLLADPTL
Mb1656|M.bovis_AF2122/97 -----IVEGTIVKVDRDEVLLDIGYKTEGVIP--------ARELSIKHDV
Rv1630|M.tuberculosis_H37Rv -----IVEGTIVKVDRDEVLLDIGYKTEGVIP--------ARELSIKHDV
MAV_3152|M.avium_104 -----IVEGTIVKVDRDEVLLDIGYKTEGVIP--------ARELSIKHDV
MLBr_01382|M.leprae_Br4923 -----IVEGTIVKVDRDEVLLDIGYKTEGVIP--------ARELSIKHDV
: .* .:*: : . :*:. *. * .* . :
MUL_3175|M.ulcerans_Agy99 APESAAAEFISDDVADAAAALEGARHILVERAAEDAELVGAIREKFWSDA
MMAR_2585|M.marinum_M APESAAAEFISDDVADAAAALEGARHILVERAAEDAELVGAIREKFWSDA
MAB_1549c|M.abscessus_ATCC_199 VPDVVAAEFLTEGVADANAALDGARHILIERAAEDAELVGATRTKFWAEG
MSMEG_2839|M.smegmatis_MC2_155 VPEEVAGEFLNENVADAAAALEGARHIIIERASEDAELVGATREKFWSEG
TH_0210|M.thermoresistible__bu VPDQVAGEYVGESVADPAAALDGARHILTDRAAEDAELVGAIREKFWVQG
Mflv_3444|M.gilvum_PYR-GCK VPEEAAAEFTSAEVADTAAALDGARHIVVERAAEDAELVGALRERFWENA
Mvan_3200|M.vanbaalenii_PYR-1 VPEQTAAEFVGEEVADVTAALEGARHIIVERAAEDAELVGGLRERFWESG
Mb1656|M.bovis_AF2122/97 DPNEVVSVGDEVEALVLTKEDKEGRLILSKKRAQYERAWGTIEALKEKDE
Rv1630|M.tuberculosis_H37Rv DPNEVVSVGDEVEALVLTKEDKEGRLILSKKRAQYERAWGTIEALKEKDE
MAV_3152|M.avium_104 DPNEVVSVGDEVEALVLTKEDKEGRLILSKKRAQYERAWGTIEALKEKDE
MLBr_01382|M.leprae_Br4923 DPNEVVSVGDEVEALVLTKEDKEGRLILSKKRAQYERAWGTIEALKEKDE
*: ... . . .* *: .: :: . * . .
MUL_3175|M.ulcerans_Agy99 SLQTAPWSADVVTTAAAQKFRDYFEFSERLHTMPSHRVLAVLRGEKEKVL
MMAR_2585|M.marinum_M SLQTAPWSADVVTTAAAQKFRDYFEFSEPLHTMPSHRVLAVLRGEKEKVL
MAB_1549c|M.abscessus_ATCC_199 ALRTAPWSEDAAKTEAAQKYRDYFEFSEPLETMPSHRVLAVMRGEKEQVL
MSMEG_2839|M.smegmatis_MC2_155 SLRTAPFSEEAAKSPAAQKFRDYFEFSEPLEQMPSHRVLAVLRGEKEQAL
TH_0210|M.thermoresistible__bu TLRTAPWSDEVATSAAAQKFRDYFEFAEPLEEMPSHRVLAVLRGEKEEVL
Mflv_3444|M.gilvum_PYR-GCK SARTRPASDAAAASEKGQKFRDYFDHTEPLSSMPSHRVLAVLRGEKEQVL
Mvan_3200|M.vanbaalenii_PYR-1 TLRTRPASEAAASAEKSQKFRDYFDYAEPLTEMPSHRVLAVLRGEKEQAL
Mb1656|M.bovis_AF2122/97 AVKGTVIEVVKGGLILDIGLRGFLP--ASLVEMRRVRDLQPYIGKEIEAK
Rv1630|M.tuberculosis_H37Rv AVKGTVIEVVKGGLILDIGLRGFLP--ASLVEMRRVRDLQPYIGKEIEAK
MAV_3152|M.avium_104 AVKGTVIEVVKGGLILDIGLRGFLP--ASLVEMRRVRDLQPYIGKEIEAK
MLBr_01382|M.leprae_Br4923 AVKGIVIEVVKGGLILDIGLRGFLP--ASLVEMRRVRDLQPYIGKEIEAK
: : . *.:: * * * * *:: :.
MUL_3175|M.ulcerans_Agy99 SLNFDCGEDSPYEAMVAATLDIDLTAKAAVTPWLAATVSFAWRIRLISSA
MMAR_2585|M.marinum_M SLNFDCGEDSPYEAMVAATLDIDLSAKAAATPWLAATVSFAWRIRLISSA
MAB_1549c|M.abscessus_ATCC_199 SLNFDGGDDAPYEHMVAQSLGVNLTANTPATPWLASTVRVAWRAKLMFSA
MSMEG_2839|M.smegmatis_MC2_155 SLTFDGGEDEAYQAMIAQALGIDMTSPAPATPWLATTVRLAWRAKLMISA
TH_0210|M.thermoresistible__bu ALTFDGGDEAGYHAMIADTLGIDLTAAAAATPWLTGTVEWAWRTKLSVSA
Mflv_3444|M.gilvum_PYR-GCK SLALDGGEDDTYRAMIASALGVDLTAGAAATPWLAATVGFAWRTRLSVSA
Mvan_3200|M.vanbaalenii_PYR-1 ALTLDGGEEDSYLAMIASALGIDLTAAAPATRWLASTVGFAWRTRLSVSA
Mb1656|M.bovis_AF2122/97 IIELDKNRNN----------------------------------------
Rv1630|M.tuberculosis_H37Rv IIELDKNRNN----------------------------------------
MAV_3152|M.avium_104 IIELDKNRNN----------------------------------------
MLBr_01382|M.leprae_Br4923 IIELDKNRNN----------------------------------------
: :* . :
MUL_3175|M.ulcerans_Agy99 TIDARVRLRQRAEVDAVSVFAKNLKDLLLAAPAGARTTLGLDPGFRTGVK
MMAR_2585|M.marinum_M TIDARVRLRQRAEVDAVSVFAKNLKDLLLAAPAGARTTLGLDPGFRTGVK
MAB_1549c|M.abscessus_ATCC_199 SVDARLKLRQRAEEDAVGVFARNLKDLLLAAPAGTRATLGLDPGFRTGVK
MSMEG_2839|M.smegmatis_MC2_155 SVDARIRLRQRAEEDAVAVFARNLKDLLLAAPAGTRTTLGLDPGFRTGVK
TH_0210|M.thermoresistible__bu KVDARIRLRQRAEEEAVAVFARNLRDLLLAAPAGSRVTLGLDPGYRTGVK
Mflv_3444|M.gilvum_PYR-GCK SVDARVRLRLRAEADAVTVFARNLKDLLLAAPAGNRTTLGLDPGFRTGVK
Mvan_3200|M.vanbaalenii_PYR-1 SVDARVRLRLRAEQDAVTVFAKNLKDLLLAAPAGNRTTMGLDPGFRTGVK
Mb1656|M.bovis_AF2122/97 VVLSRRAWLEQTQSEVRSEFLNNLQKGTIRKGVVSSIVN-----FGAFVD
Rv1630|M.tuberculosis_H37Rv VVLSRRAWLEQTQSEVRSEFLNNLQKGTIRKGVVSSIVN-----FGAFVD
MAV_3152|M.avium_104 VVLSRRAWLEQTQSEVRSEFLNQLQKGAIRKGVVSSIVN-----FGAFVD
MLBr_01382|M.leprae_Br4923 VVLSRRAWLEQTQSEVRSEFLNQLQKGAIRKGVVSSIVN-----FGAFVD
: :* ::: :. * .:*:. : . . : : *.
MUL_3175|M.ulcerans_Agy99 VAVVDGTGKVVDTCAIYPHQPQRQWDTAKATLAALIARHRVNLIAIGNGT
MMAR_2585|M.marinum_M VAVVDGTGKVVDTCAIYPHQPQRQWDTAKATLAALIARHRVDLIAIGNGT
MAB_1549c|M.abscessus_ATCC_199 VAVVDGTGKVLDTCAIYPHQPQKKWDEAKATLAAFIARHNVELIAVGNGT
MSMEG_2839|M.smegmatis_MC2_155 VAVVDGTGKVLETCAIYPHQPQKQWDQAKATLGALIARHGVELIAIGNGT
TH_0210|M.thermoresistible__bu VAVVDGTGKVLDTATIYPHQPQKQWDAAKATLAALVARHGVELIAVGNGT
Mflv_3444|M.gilvum_PYR-GCK VAVVDGTGKVLDTCAIYPHQPQKQWDTAKATLAALVARHGVELIAVGNGT
Mvan_3200|M.vanbaalenii_PYR-1 VAVVDGTGKVLDTCAIYPHQPQKQWDAAKATLAALVARHGVELIAIGNGT
Mb1656|M.bovis_AF2122/97 LGGVDGLVHVSELSWKHIDHPS-------------------EVVQVG---
Rv1630|M.tuberculosis_H37Rv LGGVDGLVHVSELSWKHIDHPS-------------------EVVQVG---
MAV_3152|M.avium_104 LGGVDGLVHVSELSWKHIDHPS-------------------EVVQVG---
MLBr_01382|M.leprae_Br4923 LGGVDGLVHVSELSWKHIDHPS-------------------EVVQVG---
:. *** :* : . : .:*. ::: :*
MUL_3175|M.ulcerans_Agy99 ASREPDALATELISDIRGAGATPPAKAVVSEAGASVYSASAYAAHELPEL
MMAR_2585|M.marinum_M ASRETDALATELISDIRGAGATPPAKAVVSEAGASVYSASAYAAHELPEL
MAB_1549c|M.abscessus_ATCC_199 ASRETDALAAELIADIKAAGAKAPTKAMVSEAGASVYSASAYAAHELPDL
MSMEG_2839|M.smegmatis_MC2_155 ASRETDALASELIADIRKSGANPPIKAMVSEAGASVYSASAYAARELPDL
TH_0210|M.thermoresistible__bu ASRETDALAAEVIADIRAAGATPPVKAMVSEAGASVYSASSYAARELPDL
Mflv_3444|M.gilvum_PYR-GCK ASRETDALATELIADIRGAGGTAPAKAMVSEAGASVYSASAYAAHELPDL
Mvan_3200|M.vanbaalenii_PYR-1 ASRETDALATELIADIRTAGANAPAKAIVSEAGASVYSASAYAAHELPDL
Mb1656|M.bovis_AF2122/97 ------DEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPG
Rv1630|M.tuberculosis_H37Rv ------DEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPG
MAV_3152|M.avium_104 ------DEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPG
MLBr_01382|M.leprae_Br4923 ------NEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPG
.: : *: . . .:: .. : * ::* ::
MUL_3175|M.ulcerans_Agy99 DVTLRGAVSIARRLQDPLAELVKIEPKSIGVGQYQHDVTPGILARSLDAV
MMAR_2585|M.marinum_M DVTLRGAVSIARRLQDPLAELVKIEPKSIGVGQYQHDVTPGILARSLDAV
MAB_1549c|M.abscessus_ATCC_199 DVTLRGAVSIARRLQDPLAELVKIEPKSIGVGQYQHDVTPGTLARSLNAV
MSMEG_2839|M.smegmatis_MC2_155 DVTVRGAVSIARRLQDPLAELVKIDPKSIGVGQYQHDVTPGSLARSLDAV
TH_0210|M.thermoresistible__bu DVTLRGAVSIARRLQDPLAELVKIEPKSIGVGQYQHDVTPGYLARSLDAV
Mflv_3444|M.gilvum_PYR-GCK DVTLRGAVSIARRLQDPLAELVKIEPKSIGVGQYQHDVTPGILAKSLGAV
Mvan_3200|M.vanbaalenii_PYR-1 DVTLRGAVSIARRLQDPLAELVKIEPKSIGVGQYQHDVTPGTLARSLGAV
Mb1656|M.bovis_AF2122/97 KVTKLVPFGAFVRVEEGIEGLVHIS-----------------ELAERHVE
Rv1630|M.tuberculosis_H37Rv KVTKLVPFGAFVRVEEGIEGLVHIS-----------------ELAERHVE
MAV_3152|M.avium_104 KVTKLVPFGAFVRVEEGIEGLVHIS-----------------ELAERHVE
MLBr_01382|M.leprae_Br4923 KVTKLVPFGAFVRVEEGIEGLVHIS-----------------ELAERHVE
.** ... *::: : **:*. . .
MUL_3175|M.ulcerans_Agy99 VEDAVNAVGVDLNTASVPLLARVSGVSESLAEAIVAYRDNTGPFHNRRAL
MMAR_2585|M.marinum_M VEDAVNAVGVDLNTASVPLLARVSGVSESLAEAIVAYRDNTGPFHNRRAL
MAB_1549c|M.abscessus_ATCC_199 VEDAVNAVGVDLNTASVPLLARVSGVTESLAESIVAHREKVGAFRSRKGL
MSMEG_2839|M.smegmatis_MC2_155 VEDAVNAVGVDLNTASVPLLARVSGVTESLAEAIVAHRESTGPFRNRKAL
TH_0210|M.thermoresistible__bu VEDAVNAVGVDLNTASVPLLSRVSGISESLAESIVAHREKTGPFRNRKAL
Mflv_3444|M.gilvum_PYR-GCK VEDAVNAVGVDLNTASVPLLSRVSGITESLAEAIVAHRDQTGRFQNRRAL
Mvan_3200|M.vanbaalenii_PYR-1 VEDAVNAVGVDLNTASVPLLARVSGITESLAEAIVAHRDKTGRFQNRRAL
Mb1656|M.bovis_AF2122/97 VPDQVVAVGDDAMVKVIDIDLERRRISLSLKQANEDYTEEFDPAKYG---
Rv1630|M.tuberculosis_H37Rv VPDQVVAVGDDAMVKVIDIDLERRRISLSLKQANEDYTEEFDPAKYG---
MAV_3152|M.avium_104 VPDQVVAVGDDAMVKVIDIDLERRRISLSLKQANEDYTEEFDPAKYG---
MLBr_01382|M.leprae_Br4923 VPDQVVAVGDDAMVKVIDIDLERRRISLSLKQANEDYIEEFDPAKYG---
* * * *** * . : : . :: ** :: : :. . :
MUL_3175|M.ulcerans_Agy99 LDVPRLGPKAFEQCAGFLRIRDGEDPLDASGVHPEAYPVVRRILDRTGVA
MMAR_2585|M.marinum_M LDVPRLGPKAFEQCAGFLRIRDGEDPLDASGVHPEAYPVVRRILDRTGVA
MAB_1549c|M.abscessus_ATCC_199 LEVPRLGPKAFEQCAGFLRIRGGDDPLDASGVHPESYPVVRKILDRSGVT
MSMEG_2839|M.smegmatis_MC2_155 LDVPRLGPKAFEQCAGFLRIRDGEDPLDASGVHPESYPVVRRILDRAGVT
TH_0210|M.thermoresistible__bu LDVPRLGPKAFEQCAGFLRIRDGDDPLDASGVHPEAYPVVRRILDRAGVT
Mflv_3444|M.gilvum_PYR-GCK LDVPRLGPKAFEQCAGFLRIRDGDDPLDASGVHPESYPVVRRILDRANVT
Mvan_3200|M.vanbaalenii_PYR-1 LDVPRLGPKAFEQCAGFLRIRDGEDPLDASGVHPESYPVVRRILDRANVT
Mb1656|M.bovis_AF2122/97 ------MADSYDEQGNYIFPEG---------------------FDAETNE
Rv1630|M.tuberculosis_H37Rv ------MADSYDEQGNYIFPEG---------------------FDAETNE
MAV_3152|M.avium_104 ------MADSYDEQGNYIFPEG---------------------FDPETNE
MLBr_01382|M.leprae_Br4923 ------MADSYDEQGNYIFPEG---------------------FDPDSNE
..:::: ..:: .. :*
MUL_3175|M.ulcerans_Agy99 LAEIIGDERSLRSLRPADFADDRFGVPTVTDILAELEKPGRDPRPAFITA
MMAR_2585|M.marinum_M LAEIIGDERSLRSLRPADFADDRFGVPTVTDILAELEKPGRDPRPAFITA
MAB_1549c|M.abscessus_ATCC_199 LAELIGNERSLRSLKPADFADDRFGIPTVTDILGELEKPGRDPRPAFTTA
MSMEG_2839|M.smegmatis_MC2_155 LAELIGDTRTLRTLKPAEFADDRFGVPTVTDIIGELEKPGRDPRPAFSTA
TH_0210|M.thermoresistible__bu LAELIGNEKTLRALRPADFADDQFGIPTVTDILAELEKPGRDPRPAFATA
Mflv_3444|M.gilvum_PYR-GCK LTEIIGDERALRALKPADFADDTFGIPTVTDILGELEKPGRDPRPAFTTA
Mvan_3200|M.vanbaalenii_PYR-1 LAEIIGNERTLRALRPADFADDRFGIPTVTDILGELEKPGRDPRPAFTTA
Mb1656|M.bovis_AF2122/97 WLEGFEKQRAEWEARYAEAERRHKMHTAQMEKFAAAETAGRGADDQSSAS
Rv1630|M.tuberculosis_H37Rv WLEGFEKQRAEWEARYAEAERRHKMHTAQMEKFAAAEAAGRGADDQSSAS
MAV_3152|M.avium_104 WLEGFEKQRAEWEARYAEAERRHKMHTAQMEKFAAAEAAGHAGGEQSPGN
MLBr_01382|M.leprae_Br4923 WLEGFDTQRAEWEARYAEAERRYKMHTIQMEKFAATEEAGHGSSEQPPAS
* : :: : *: . : :. * .*:
MUL_3175|M.ulcerans_Agy99 TIAAGVEKVADLKVGMVLEGVVTNVAAFGAFVDVGVHQDGLVHVAAMAER
MMAR_2585|M.marinum_M TFAAGVEKVADLKVGMVLEGVVTNVAAFGAFVDVGVHQDGLVHVSAMAER
MAB_1549c|M.abscessus_ATCC_199 TFAAGVEKVSDLKAGMVLEGVVTNVAAFGAFVDVGVHQDGLVHVSAMSDR
MSMEG_2839|M.smegmatis_MC2_155 TFAAGVEKVADLKPGMILEGVVTNVAAFGAFVDVGVHQDGLVHVSAMADR
TH_0210|M.thermoresistible__bu TFTDGVDTIADLKVGMVLEGVVTNVAAFGAFVDIGVHQDGLVHVSAMADR
Mflv_3444|M.gilvum_PYR-GCK TFAAGVEKVADLKAGMILEGVVTNVAAFGAFVDVGVHQDGLVHVSAMADR
Mvan_3200|M.vanbaalenii_PYR-1 TFAAGVEKVADLKVGMILEGVVTNVAAFGAFVDVGVHQDGLVHVSAMADR
Mb1656|M.bovis_AF2122/97 SAPSEKTAGGSLASDAQLAALREKLAGSA---------------------
Rv1630|M.tuberculosis_H37Rv SAPSEKTAGGSLASDAQLAALREKLAGSA---------------------
MAV_3152|M.avium_104 GAPAEK-AGGSLASDAQLAALREKLAGNA---------------------
MLBr_01382|M.leprae_Br4923 STPSAKATGGSLASDAQLAALREKLAGSA---------------------
. ..* . * .: ::*. .
MUL_3175|M.ulcerans_Agy99 FVSDPHEVVRSGQVVRVKVVDVDVERQRVGLSLRLNDEPQR-----DRAN
MMAR_2585|M.marinum_M FVSDPHEVVRSGQVVRVKVVDVDVERQRIGLSLRLNDEPQR-----DRAN
MAB_1549c|M.abscessus_ATCC_199 YVSDPHEVVKSGQVVRVKVTEVDIPRQRIGLTLRLNDDPQQ-GKRSERSS
MSMEG_2839|M.smegmatis_MC2_155 FVSDPHEVVKSGQVVKVKVVDVDVDRQRIGLSLRLNDEPDK-GKRNDRRD
TH_0210|M.thermoresistible__bu FVSDPHDIVRSGQVVRVKVVDVDVERQRIGLSLRLDDDPHQSGKRSGNRG
Mflv_3444|M.gilvum_PYR-GCK YVSDPHEVVRSGQVVRVKVVDVDVERQRIGLSMRLNDETTP------QRG
Mvan_3200|M.vanbaalenii_PYR-1 YISDPHEVVRSGQVVRVKVVDVDVDRQRIGLSLRLKDDVKP------ERG
Mb1656|M.bovis_AF2122/97 --------------------------------------------------
Rv1630|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3152|M.avium_104 --------------------------------------------------
MLBr_01382|M.leprae_Br4923 --------------------------------------------------
MUL_3175|M.ulcerans_Agy99 RADRAPKPRGGADSKLRR----DQRNPHHAKCAGRRDSSSSGSSMAQALR
MMAR_2585|M.marinum_M RADRAPKPRGGADSKLRR----DQRNPHHAKGAGRRDSSSSGSSMAQALR
MAB_1549c|M.abscessus_ATCC_199 APRRDDRNRD--DRNQG------RRNQNRDKNSGRREESRPTGSMADALR
MSMEG_2839|M.smegmatis_MC2_155 GGQNRNQNRGGGQRNKGGNSGGNKGGNNRGNKGGRRDQ--PMGSMAEALR
TH_0210|M.thermoresistible__bu AKGEAPKGGNGRRPDRG-----DRRNQNRSRGGN-----QPNGAMAQALR
Mflv_3444|M.gilvum_PYR-GCK P-RRNERPRT------------PQRKPQRPNNTGGRETGKAGGSMARALR
Mvan_3200|M.vanbaalenii_PYR-1 GGRRGDRPAN------------PKRNPQRANNSGRREAGSGGGSMAQALR
Mb1656|M.bovis_AF2122/97 --------------------------------------------------
Rv1630|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3152|M.avium_104 --------------------------------------------------
MLBr_01382|M.leprae_Br4923 --------------------------------------------------
MUL_3175|M.ulcerans_Agy99 DAGFGK
MMAR_2585|M.marinum_M DAGFGK
MAB_1549c|M.abscessus_ATCC_199 NAGFGR
MSMEG_2839|M.smegmatis_MC2_155 NAGFGR
TH_0210|M.thermoresistible__bu DAGFGR
Mflv_3444|M.gilvum_PYR-GCK DAGFGR
Mvan_3200|M.vanbaalenii_PYR-1 EAGFGR
Mb1656|M.bovis_AF2122/97 ------
Rv1630|M.tuberculosis_H37Rv ------
MAV_3152|M.avium_104 ------
MLBr_01382|M.leprae_Br4923 ------