For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQAISLAAVLSAFSAGIAIATRLLMPGPDGRPT AGIIDAEGRLGWPLWAAYVSVFALTVLVGVRLVVAVVRVAIATRRRRARHRMVVDLVGVGHEATLAQPCA RTRDLRVLHVAQPLAYCLPGVRSRVVVSEGALTTLSDEEVAAILTHERAHLRARHDLVLEAFTAFHEAFP RLVRSANALRAAQLLVELLADDPAVRAAGRTPLARALVACASGRAPLGALAAGGPSTLVRVQRLSGRGNS LALAAAAYAAAAVVLVVPTVALAVPWLTELQRLFDL
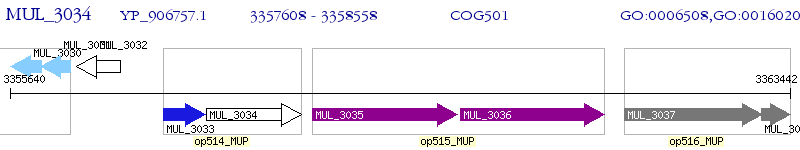
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3034 | - | - | 100% (316) | hypothetical protein MUL_3034 |
| M. ulcerans Agy99 | MUL_4135 | - | 6e-22 | 31.80% (283) | peptidase M48 like- protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1876c | - | 1e-151 | 87.26% (314) | hypothetical protein Mb1876c |
| M. gilvum PYR-GCK | Mflv_3358 | - | 1e-125 | 73.57% (314) | peptidase M48, Ste24p |
| M. tuberculosis H37Rv | Rv1845c | - | 1e-151 | 87.26% (314) | hypothetical protein Rv1845c |
| M. leprae Br4923 | MLBr_02064 | - | 1e-145 | 82.91% (316) | integral membrane protein |
| M. abscessus ATCC 19977 | MAB_2414c | - | 1e-119 | 68.42% (323) | hypothetical protein MAB_2414c |
| M. marinum M | MMAR_2719 | - | 1e-172 | 99.68% (316) | hypothetical protein MMAR_2719 |
| M. avium 104 | MAV_2870 | - | 1e-147 | 84.18% (316) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_3631 | - | 1e-121 | 70.38% (314) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3088 | - | 1e-125 | 74.84% (314) | peptidase M48, Ste24p |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3358|M.gilvum_PYR-GCK MSALAFSLLALLLSGPVPAMLARAAWPLRAPRAAIVLWQSIAAAAVLSAF
Mvan_3088|M.vanbaalenii_PYR-1 MSALAFTIVALLLSGPVPALLARASWPLRAPRAAIVLWQSIALAAVLSAF
MSMEG_3631|M.smegmatis_MC2_155 MSALAFTLVALALVGPVPAMLARASWPLRAPRAAIVLWQSIALAAVLSAF
MUL_3034|M.ulcerans_Agy99 MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQAISLAAVLSAF
MMAR_2719|M.marinum_M MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQAISLAAVLSAF
Mb1876c|M.bovis_AF2122/97 MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQAIALAAVLSSF
Rv1845c|M.tuberculosis_H37Rv MSALAFTILAVLLAGPTPALLARATWPLRAPRAAMVLWQAIALAAVLSSF
MLBr_02064|M.leprae_Br4923 MSALAFTILAVLLVGPTPTLVARSTWPLRAPRAAMVLWQTIALAAALSTF
MAV_2870|M.avium_104 MSALAFTILAVLLTGPVPAMLARARWPLRAPRAAMVLWQAVALAAVLSAF
MAB_2414c|M.abscessus_ATCC_199 MSALAFAILALLLTGPVPALLARARWPYRAPRAAMVLWQAIAVAAVLSAF
******:::*: * **.*:::**: ** ******:****::: **.**:*
Mflv_3358|M.gilvum_PYR-GCK SAGIAIASRLFVPGPDGRPTSTIVGEIAVLGWPLWTAYVVVFLLTLVIGA
Mvan_3088|M.vanbaalenii_PYR-1 SAGIAIASRLFVPGPDGRPTATIVSEIEVLGWPLWTAYVVVFVLTLMIGA
MSMEG_3631|M.smegmatis_MC2_155 SAGIAIASRLFVPGADGRPTATITSEIDVLGWPLWLLYVVVFSLTLFIGA
MUL_3034|M.ulcerans_Agy99 SAGIAIATRLLMPGPDGRPTAGIIDAEGRLGWPLWAAYVSVFALTVLVGV
MMAR_2719|M.marinum_M SAGIAIATRLLMPGPDGRPTAGIIDAEGRLGWPLWAAYVSVFALTVLVGV
Mb1876c|M.bovis_AF2122/97 SAGIAIASRLLMPGPDGRPTTSFVGAAGRLGWPLWAAYITVFALTVLVGA
Rv1845c|M.tuberculosis_H37Rv SAGIAIASRLLMPGPDGRPTTSFVGAAGRLGWPLWAAYITVFALTVLVGA
MLBr_02064|M.leprae_Br4923 SAGIAIASRVLMPGPDGRLTASVIGAAGRLGWPLWAAYVAAFALTVLVGA
MAV_2870|M.avium_104 SAGIAIATRVLVPGPDGRPTTSILGAEGRLGWPLWTAYIGVFALTVLVGA
MAB_2414c|M.abscessus_ATCC_199 SSGLAIASRLLVPGPDGHPTATITGEIDRLGWGLWLLYVTVFAITILIGV
*:*:***:*:::**.**: *: . . *** ** *: .* :*:.:*.
Mflv_3358|M.gilvum_PYR-GCK RLVVSVLNVAIATRRRRAHHRMVVDLVG-----AHHSR-----TRSRSSL
Mvan_3088|M.vanbaalenii_PYR-1 RLIVSVLQVAIATRRRRAHHRMVVDLVG-----SHDHR-----HRSGAGL
MSMEG_3631|M.smegmatis_MC2_155 RLCVAIVQVGVATRRRRAHHRMMVDLLSKSRDAVPAHL-----RTPASGL
MUL_3034|M.ulcerans_Agy99 RLVVAVVRVAIATRRRRARHRMVVDLVGVGHEATLAQP-----CARTRDL
MMAR_2719|M.marinum_M RLVVAVVRVAIATRRRRARHRMVVDLVGVGHEATLAQP-----CARTRDL
Mb1876c|M.bovis_AF2122/97 RLAVAVVRVATATRRRRAHHRMVVDLVGVGHNGALAQP-----CARARDL
Rv1845c|M.tuberculosis_H37Rv RLAVAVVRVATATRRRRAHHRMVVDLVGVGHNGALAQP-----CARARDL
MLBr_02064|M.leprae_Br4923 RLIVAIVRVAIATRRRRAHHRMVVDLVGVGHNAALAQP-----CARARDL
MAV_2870|M.avium_104 RLMVAVVRVAIANRRRRAHHRMVVDLVGMGHGAALSQP-----CSRTRDL
MAB_2414c|M.abscessus_ATCC_199 RLMVSVVRVGVRTRRRRARHRAIVDLLDHKRYCENWRGGYDINRRRDHDL
** *:::.*. .*****:** :***:. .*
Mflv_3358|M.gilvum_PYR-GCK RVLDVDEPLAYCLPGVRSRVVVSEGALKTLADNEMSAILEHEHAHLRARH
Mvan_3088|M.vanbaalenii_PYR-1 RILDVAEPLAYCLPGVRSRVVVSEGALKALSDNEMAAILEHEHAHLRARH
MSMEG_3631|M.smegmatis_MC2_155 RILDVAQPLAYCLPGVRSRVVVSEGTLDTLSPDEVDAILTHERAHLRARH
MUL_3034|M.ulcerans_Agy99 RVLHVAQPLAYCLPGVRSRVVVSEGALTTLSDEEVAAILTHERAHLRARH
MMAR_2719|M.marinum_M RVLHVAQPLAYCLPGVRSRVVVSEGALTTLSDEEVAAILTHERAHLRARH
Mb1876c|M.bovis_AF2122/97 RVLDVAQPLAYCLPGVRSRVVVSEGTLTALADAEVAAILTHERAHLRARH
Rv1845c|M.tuberculosis_H37Rv RVLDVAQPLAYCLPGVRSRVVVSEGTLTALADAEVAAILTHERAHLRARH
MLBr_02064|M.leprae_Br4923 RVLEVAQPLAYCLPGVRSRVVVSEGALTKLNDTEVTAILTHERAHLRARH
MAV_2870|M.avium_104 RVLDVPQPLAYCLPGVRSRVVVSEGTLSTLADAEVSAILTHERAHLRARH
MAB_2414c|M.abscessus_ATCC_199 RVLEVDEPLAYCLPGVRSRVVVSEGTLSTLGHNEVTAIVAHERAHLRARH
*:*.* :******************:* * *: **: **:*******
Mflv_3358|M.gilvum_PYR-GCK DLVLEMFTAVHAAFPRLVRSGHALNAVRLLIELLADDAAVRAEGPTPLAR
Mvan_3088|M.vanbaalenii_PYR-1 DLVLEMFTAVHAAFPRLVRSGHALNAVRLLIELLADDAAVRAEGPTPLAR
MSMEG_3631|M.smegmatis_MC2_155 DLVLEMFTAVHAAFPRFVRSASALDAVRLLIELLADDAAVRIAGPTPLAR
MUL_3034|M.ulcerans_Agy99 DLVLEAFTAFHEAFPRLVRSANALRAAQLLVELLADDPAVRAAGRTPLAR
MMAR_2719|M.marinum_M DLVLEAFTAFHEAFPRLVRSANALRAAQLLVELLADDAAVRAAGRTPLAR
Mb1876c|M.bovis_AF2122/97 DLVLEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRTPLAR
Rv1845c|M.tuberculosis_H37Rv DLVLEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRTPLAR
MLBr_02064|M.leprae_Br4923 DLVLEAFTAVHAAFPRLVRSSAALSAVRLLVELLADDAAVRAAGRTPLAR
MAV_2870|M.avium_104 DLVLEAFTAVHAAFPRLVRSANALGAVQLLVELLADDAAVRAAGRAPLAR
MAB_2414c|M.abscessus_ATCC_199 DLVLEAFTAVHDAFPVIVRSKSALDAVKLLVELLADDAAVRTAGPTPLAR
***** ***.* *** :*** ** *.:**:******.*** * :****
Mflv_3358|M.gilvum_PYR-GCK ALVACASGRAPRGALAAGGPTTVVRVRRLGGKPNSRMLAAGAYAAAAAVL
Mvan_3088|M.vanbaalenii_PYR-1 ALVACASGRAPRGALAAGGPTTVVRVRRLGGAPNSRILAAGAYITAAAVL
MSMEG_3631|M.smegmatis_MC2_155 ALVTCAAGRTPSGALAAGGPTTVIRVRRLGGKPNSLTLSVIAYLAAVAIL
MUL_3034|M.ulcerans_Agy99 ALVACASGRAPLGALAAGGPSTLVRVQRLSGRGNSLALAAAAYAAAAVVL
MMAR_2719|M.marinum_M ALVACASGRAPLGALAAGGPSTLVRVQRLSGRGNSLALAAAAYAAAAVVL
Mb1876c|M.bovis_AF2122/97 ALVACASGRAPSGALAVGGPSTVLRVRRLSGRGNSAVLSAAAYLAAAAVL
Rv1845c|M.tuberculosis_H37Rv ALVACASGRAPSGALAVGGPSTVLRVRRLSGRGNSAVLSAAAYLAAAAVL
MLBr_02064|M.leprae_Br4923 ALVACASGQAPSGALAAGGNTTVLRVRRLSGRSNSAVVSAAAYLAAAIVL
MAV_2870|M.avium_104 ALVACASGRAPSGALAAGGPSTVVRVRRLGGRGNSPLLSAAAYLAAAAVF
MAB_2414c|M.abscessus_ATCC_199 ALVACAGGPTPVGAMAAGGPTTLIRVRRLGGSGNSLLLSCSAYTAAAAIL
***:**.* :* **:*.** :*::**:**.* ** :: ** :*. ::
Mflv_3358|M.gilvum_PYR-GCK VVPTVAFAVPWLNELHRLFLV--
Mvan_3088|M.vanbaalenii_PYR-1 VVPTVALAVPWLTELQRLFSA--
MSMEG_3631|M.smegmatis_MC2_155 VVPTVAVAVPWLTELHRLFSGLT
MUL_3034|M.ulcerans_Agy99 VVPTVALAVPWLTELQRLFDL--
MMAR_2719|M.marinum_M VVPTVALAVPWLTELQRLFDL--
Mb1876c|M.bovis_AF2122/97 VVPTVALAVPWLTQLQRLFIA--
Rv1845c|M.tuberculosis_H37Rv VVPTVALAVPWLTQLQRLFIA--
MLBr_02064|M.leprae_Br4923 LVPTVALAVPWLTELQRLFNI--
MAV_2870|M.avium_104 VVPTVALAVPWLTELERLLNL--
MAB_2414c|M.abscessus_ATCC_199 VVPTLAVVIPWFTELQRLFSK--
:***:*..:**:.:*.**: