For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FGCLGELAIAQRCRREVNRLCCEVGGRIGWGRRWPDVPVGNGCILPASQRQPTSRVQAMTHHPKRLLVTV VGAFAAGVLAPATAHADIVGFTSPSGNIGCMIDSSSVRCDVRDRTWSPPPKPADCESMMNYGQGIVLTAG QPARFVCAGDTALGGGPALAYGDKITKGSLQCESETSGVTCRDIGDGGGFSISRDGYQLS*
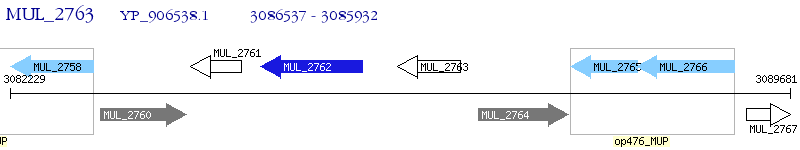
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2763 | - | - | 100% (201) | hypothetical protein MUL_2763 |
| M. ulcerans Agy99 | MUL_2262 | lppI | 3e-06 | 25.64% (195) | lipoprotein LppI |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2072 | lppI | 7e-07 | 32.71% (107) | lipoprotein lppI |
| M. gilvum PYR-GCK | Mflv_3890 | - | 2e-36 | 54.10% (122) | hypothetical protein Mflv_3890 |
| M. tuberculosis H37Rv | Rv2046 | lppI | 7e-07 | 32.71% (107) | lipoprotein lppI |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3181 | - | 7e-06 | 31.09% (119) | lipoprotein LppI |
| M. marinum M | MMAR_3530 | - | 1e-80 | 98.59% (142) | hypothetical protein MMAR_3530 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2525 | - | 6e-36 | 56.39% (133) | hypothetical protein Mvan_2525 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2763|M.ulcerans_Agy99 -------MFGCLGELAIAQRCRREVNRLCCEVGGRIGWGRRWPDVPVGNG
MMAR_3530|M.marinum_M --------------------------------------------------
Mflv_3890|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2525|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2072|M.bovis_AF2122/97 MRIAAL-----VAVSLLIAGCPREVGGDVGQSQTIAPPAPAPSAAPSTP-
Rv2046|M.tuberculosis_H37Rv MRIAAL-----VAVSLLIAGCSREVGGDVGQSQTIAPPAPAPSAAPSTP-
MAB_3181|M.abscessus_ATCC_1997 MRALLLGGLTLLGAMLLVAGCSKGATVSPTVPSSSQPPSSSSTSKPITTT
MUL_2763|M.ulcerans_Agy99 ------CILPASQRQPTSRVQAMTHHPKRLLVTVVGAFAAGVLAPATAHA
MMAR_3530|M.marinum_M ----------------------MTHHPKRLLVTVVGAFAAGVLAPATAHA
Mflv_3890|M.gilvum_PYR-GCK -----------------------------------------MLTSPTATA
Mvan_2525|M.vanbaalenii_PYR-1 --------------------------MKQVVVAAALAGTALLGPAPVAGA
Mb2072|M.bovis_AF2122/97 -----------PAAGAPITTIVSWIEAGHPVDPAAYHVAT-RDGVTTQLG
Rv2046|M.tuberculosis_H37Rv -----------PAAGAPITTIVSWIEAGHPVDPAAYHVAT-RDGVTTQLG
MAB_3181|M.abscessus_ATCC_1997 TTTTAPVDLPEPVAGAPYDAVAQWISKGAPVDVTNFHAATSQDGQKTDLG
. .
MUL_2763|M.ulcerans_Agy99 DIVGFTSPSGNIGCMIDS----SSVRCDVRDRTWSPPPKPADCESMMNYG
MMAR_3530|M.marinum_M DIVGFTSPSGNIGCMIDS----SSVRCDVRDRTWSPPPRPADCESMMDYG
Mflv_3890|M.gilvum_PYR-GCK DLVGFTSPSGNIGCILTD----EMVRCDIASRSWDPPPRPADCPDVTGYG
Mvan_2525|M.vanbaalenii_PYR-1 DIVGFTSPSGNIGCMLDD----DMVRCDIAARDWAPPPRPADCPDFTGFG
Mb2072|M.bovis_AF2122/97 DDVAFSASSGTVACMTDARHTSGTLACLVRLANPPPRPETAYGEWKGGWV
Rv2046|M.tuberculosis_H37Rv DDVAFSASSGTVACMTDARHTSGTLACLVRLANPPPRPETAYGEWKGGWV
MAB_3181|M.abscessus_ATCC_1997 DNVAFKTPSGKTRCMTDN-IQPGTLSCLVNLTNPPKRPAGTEGNWVGGWT
* *.*.:.**. *: : * : * : .:
MUL_2763|M.ulcerans_Agy99 QGIVLTAGQPARFVCAGDTALGGGPALAYGDKITKGSLQCESETSGVTCR
MMAR_3530|M.marinum_M QGIVLTAGQPARFVCAGDTALGGGPALAYGDKITKGSLQCESETSGVTCR
Mflv_3890|M.gilvum_PYR-GCK QGITLSPTGPAAFVCAGDTTLGSGPALPFGQYEAGGGMSCNSEPSGMRCS
Mvan_2525|M.vanbaalenii_PYR-1 QGIYLESTGHAAFVCAGDTAMSSGPPLDYGQVRAAGGISCTSEPSGMRCA
Mb2072|M.bovis_AF2122/97 DFDGIHLQVGSARADPGPFVYGNGPELANGDTLSIGDYRCRSYQAGLFCV
Rv2046|M.tuberculosis_H37Rv DFDGIHLQVGSARADPGPFVYGNGPELANGDTLSIGDYRCRSYQAGLFCV
MAB_3181|M.abscessus_ATCC_1997 DFSGRQVTVGGLHGDPGPFQSGDGNPLDYGGRLSFGDFSCRSETSGLFCA
: . .* ..* * * : *. * * :*: *
MUL_2763|M.ulcerans_Agy99 DIGDGGGFSISRDGYQLS------------------
MMAR_3530|M.marinum_M DIGDGGGFSISRDGYQLS------------------
Mflv_3890|M.gilvum_PYR-GCK NS-DGHGFVLAREAYQVF------------------
Mvan_2525|M.vanbaalenii_PYR-1 NS-DGHGFVISRQAYQLF------------------
Mb2072|M.bovis_AF2122/97 NYAHQSAVRFASAGIEPFGCLKPAPPPDGVGVAFGC
Rv2046|M.tuberculosis_H37Rv NYAHQSAVRFASAGIEPFGCLKPAPPPDGVGVAFGC
MAB_3181|M.abscessus_ATCC_1997 NTKAKSAIRISDAGIEPFGCLQEATPSEGNGREFSC
: .. :: . :