For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPLPPDPNPTLSAYAHPERLVTADWLSAHMGVPGLAIVESDENVLLYDIGHIPGAVKIDWHTDLNDPRVR DYINGAQFAELMDRKGISRDDTVVIYGDKSSWWAAYALWVFTLFGHPDVRSLNGGRNLWIAEGRDTSLTV PAKTSAGYPIVERNDAPIRAYKDDVLATLGTQPLIDVRSPDEYTGKRTHMPDYPEEGVLRGGHIPTAQSI PWAKAVDESGRFRGRAELDELYGFLDPDDKTIVYCRIGERSSHTWFVLTHLLGKPGVRNYDGSWTEWGNT VRVPIVGGESPGSADGLASPR
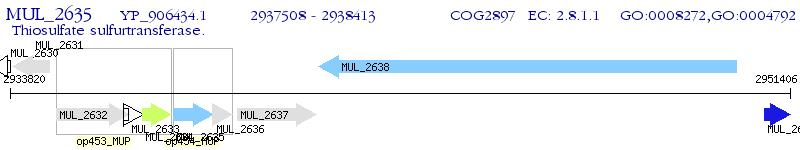
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2635 | sseA | - | 100% (301) | thiosulfate sulfurtransferase SseA |
| M. ulcerans Agy99 | MUL_0432 | cysA2 | 2e-72 | 47.10% (276) | thiosulfate sulfurtransferase, CysA2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3311 | sseA | 1e-152 | 83.90% (292) | thiosulfate sulfurtransferase SseA |
| M. gilvum PYR-GCK | Mflv_4780 | - | 1e-148 | 80.00% (300) | rhodanese domain-containing protein |
| M. tuberculosis H37Rv | Rv3283 | sseA | 1e-153 | 84.25% (292) | thiosulfate sulfurtransferase SseA |
| M. leprae Br4923 | MLBr_00728 | sseA | 1e-149 | 82.25% (293) | putative thiosulfate sulfurtransferase |
| M. abscessus ATCC 19977 | MAB_3641 | - | 1e-139 | 77.89% (294) | putative thiosulfate sulfurtransferase |
| M. marinum M | MMAR_1253 | sseA | 1e-180 | 98.34% (301) | thiosulfate sulfurtransferase SseA |
| M. avium 104 | MAV_4253 | - | 1e-160 | 87.41% (294) | putative thiosulfate sulfurtransferase |
| M. smegmatis MC2 155 | MSMEG_1809 | - | 1e-149 | 80.95% (294) | putative thiosulfate sulfurtransferase |
| M. thermoresistible (build 8) | TH_0246 | sseA | 1e-146 | 78.38% (296) | PROBABLE THIOSULFATE SULFURTRANSFERASE SSEA (RHODANESE) |
| M. vanbaalenii PYR-1 | Mvan_1669 | - | 1e-152 | 82.99% (294) | rhodanese domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4780|M.gilvum_PYR-GCK ------------------MPLPADPSPALQSYAHPERLVTADWLSGNLGR
Mvan_1669|M.vanbaalenii_PYR-1 MDTPSAKTTPHLPGRLVHVPLPADPSPALQSYAHPERLVTADWLSGNLGR
MSMEG_1809|M.smegmatis_MC2_155 ------------------MSLPADPSPTLAEYAHPERLVTADWLASNLGR
TH_0246|M.thermoresistible__bu ------------------VPLPPDPSPELKQYAHPERLVTADWLASNLGR
MUL_2635|M.ulcerans_Agy99 ------------------MPLPPDPNPTLSAYAHPERLVTADWLSAHMGV
MMAR_1253|M.marinum_M ------------------MPLPPDPNPALSAYAHPERLVTADWLSAHMGV
Mb3311|M.bovis_AF2122/97 ------------------MPLPADPSPTLSAYAHPERLVTADWLSAHMGA
Rv3283|M.tuberculosis_H37Rv ------------------MPLPADPSPTLSAYAHPERLVTADWLSAHMGA
MLBr_00728|M.leprae_Br4923 ------------------MPLPTDPSPSLSAYAHPERLVTGDWLYFHLGK
MAV_4253|M.avium_104 ------------------MPLPPDPHPSLQEYAHPERLVTADWLSANLGA
MAB_3641|M.abscessus_ATCC_1997 ------------------MTLPADPSPEFAAYAHPERLVSADWLSGHLGT
:.**.** * : ********:.*** ::*
Mflv_4780|M.gilvum_PYR-GCK PGLAIVESDEDVLLYDTGHIPGAVKIDWHTDLNDPNVRDYITGEQFADLM
Mvan_1669|M.vanbaalenii_PYR-1 PGLAIVESDEDVLLYDTGHIPGAVKIDWHIDLNDPHVRDYITGEQFADLM
MSMEG_1809|M.smegmatis_MC2_155 PGLAIVESDEDVLLYDTGHIPGAVKIDWHTDLNDPHVRDYINGEQFAALM
TH_0246|M.thermoresistible__bu PGLVIVESDEDVLLYDTGHIPGAVKIDWHTDLNDPAVRDYINGEQFAALM
MUL_2635|M.ulcerans_Agy99 PGLAIVESDENVLLYDIGHIPGAVKIDWHTDLNDPRVRDYINGAQFAELM
MMAR_1253|M.marinum_M PGLAIVESDEDVLLYDIGHIPGAVKIDWHTDLNDPRVRDYINGAQFAELM
Mb3311|M.bovis_AF2122/97 PGLAIVESDEDVLLYDVGHIPGAVKIDWHTDLNDPRVRDYINGEQFAELM
Rv3283|M.tuberculosis_H37Rv PGLAIVESDEDVLLYDVGHIPGAVKIDWHTDLNDPRVRDYINGEQFAELM
MLBr_00728|M.leprae_Br4923 PGLAIVESDENVLLYDVGHIPGAVKVDWHTDLNDPKVRDYITGEQFADLM
MAV_4253|M.avium_104 PGLVIVESDEDVLLYDVGHIPGAVKIDWHTDLNDPRVRDYIDGARFAELM
MAB_3641|M.abscessus_ATCC_1997 PGLSIVESDEDVLLYDIGHVPGAVKIDWHVDLNDSTVRDYIDGAQFAELM
*** ******:***** **:*****:*** ****. ***** * :** **
Mflv_4780|M.gilvum_PYR-GCK NRKGISRDDTVVIYGDKSNWWAAYALWVFTLFGHPDVRLLDGGRSLWISD
Mvan_1669|M.vanbaalenii_PYR-1 NRKGISRDDTVVIYGDKSNWWAAYALWVFTLFGHPDVRLLDGGRNLWISD
MSMEG_1809|M.smegmatis_MC2_155 DRKGISRDDTVVIYGDKSNWWAAYALWVFTLFGHPDVRLLDGGRDLWVSH
TH_0246|M.thermoresistible__bu DRKGVTRDDTVVIYGDKSNWWAAYAMWVFHLFGHPDVRLLDGGRDLWVST
MUL_2635|M.ulcerans_Agy99 DRKGISRDDTVVIYGDKSSWWAAYALWVFTLFGHPDVRSLNGGRNLWIAE
MMAR_1253|M.marinum_M DRKGISRDDTVVIYGDKSNWWAAYALWVFTLFGHPDVRLLNGGRDLWIAE
Mb3311|M.bovis_AF2122/97 DRKGIARDDTVVIYGDKSNWWAAYALWVFTLFGHADVRLLNGGRDLWLAE
Rv3283|M.tuberculosis_H37Rv DRKGIARDDTVVIYGDKSNWWAAYALWVFTLFGHADVRLLNGGRDLWLAE
MLBr_00728|M.leprae_Br4923 NRKGIARDDTVVIYGDKSNWWAAYALWVFTLFGHPDVRLLNGGRDLWLAE
MAV_4253|M.avium_104 DRKGISRDDTVVIYGDKSNWWAAYALWVFTLFGHPDVRLLNGGRDLWLAE
MAB_3641|M.abscessus_ATCC_1997 NRKGIRREDTVVIYGDKSNWWAAYALWVFTLFGHPDVRLLDGGRDLWISE
:***: *:**********.******:*** ****.*** *:***.**::
Mflv_4780|M.gilvum_PYR-GCK GRDTTLDVPTKQTTGYPVVERNDAPIRAFKEDVLAVLGTEPLIDVRSPQE
Mvan_1669|M.vanbaalenii_PYR-1 GRDTTLDVPSKQTTGYPVVERNDAPIRAYKDDVLAILGTQPLIDVRSPQE
MSMEG_1809|M.smegmatis_MC2_155 GRDTTLDVPTRQSSGYPVVERNDAPIRAFKEDVLEILGKQPLIDVRSPQE
TH_0246|M.thermoresistible__bu GRDTTLEVPTRQTSGYPVVERNDAPIRAFKDDVLRVLGKEPLIDVRSPQE
MUL_2635|M.ulcerans_Agy99 GRDTSLTVPAKTSAGYPIVERNDAPIRAYKDDVLATLGTQPLIDVRSPDE
MMAR_1253|M.marinum_M GRDTSLTVPAKTSAGYPIVERNDAPIRAYKDDVLATLGTQPLIDVRSPDE
Mb3311|M.bovis_AF2122/97 RRETTLDVPTKTCTGYPVVQRNDAPIRAFRDDVLAILDAQPLIDVRSPEE
Rv3283|M.tuberculosis_H37Rv RRETTLDVPTKTCTGYPVVQRNDAPIRAFRDDVLAILGAQPLIDVRSPEE
MLBr_00728|M.leprae_Br4923 RRDTSLAVPNKTSTSYPVVNRNDAPIRAFKDDVLAILGTQPLIDVRSLDE
MAV_4253|M.avium_104 RRETTLDVPTKTSTGYPVVTRNDAPIRAFKDDVLAILGSQPLIDVRSPDE
MAB_3641|M.abscessus_ATCC_1997 GRDTTLDVPSKHTEGYPVVERNDAPIRAYADDVLAHLGHGPLIDVRSPAE
*:*:* ** : .**:* ********: :*** *. ******* *
Mflv_4780|M.gilvum_PYR-GCK YTGERTHMPDYPEEGALRGGHIPTAVSIPWAKAALDNGKFRSRAELDELY
Mvan_1669|M.vanbaalenii_PYR-1 YTGERTHMPDYPEEGALRGGHIPTAVSIPWAKAALDNGKFRSRAELEELY
MSMEG_1809|M.smegmatis_MC2_155 YTGERTHMPDYPEEGALRGGHIPTAKSIPWGKAAKDNGQFRSRAELEELY
TH_0246|M.thermoresistible__bu YTGERTHMPDYPEEGALRGGHIPTAVSVPWSKAAYDDGRFRARAELEEIY
MUL_2635|M.ulcerans_Agy99 YTGKRTHMPDYPEEGVLRGGHIPTAQSIPWAKAVDESGRFRGRAELDELY
MMAR_1253|M.marinum_M YTGKRTHMPDYPEEGVLRGGHIPTAQSIPWAKAVDESGRFRGRAELDELY
Mb3311|M.bovis_AF2122/97 YTGKRTHMPDYPEEGALRAGHIPTAVHIPWGKAADESGRFRSREELERLY
Rv3283|M.tuberculosis_H37Rv YTGKRTHMPDYPEEGALRAGHIPTAVHIPWGKAADESGRFRSREELERLY
MLBr_00728|M.leprae_Br4923 YTGKCTEMPDSPEESVLRAGHIPTARSIPWEMTVDKSGRFRSSEELERLY
MAV_4253|M.avium_104 YTGKRTHMPEYPEEGVLRGGHIPTARSIPWAKAVDESGRFRSRAELEELY
MAB_3641|M.abscessus_ATCC_1997 YTGERTHMPDYPEEGALRGGHIPTAVSIPWAKAAAEDSRFRRRGELDEIY
***: *.**: ***..**.****** :** :. ...:** **:.:*
Mflv_4780|M.gilvum_PYR-GCK GFLSEHDNPETVVYCRIGERSSHTWFVLTHLLGMPGVRNYDGSWTEWGNA
Mvan_1669|M.vanbaalenii_PYR-1 GFLKPED--KTVVYCRIGERSSHTWFVLTHLLGKPGVRNYDGSWTEWGNA
MSMEG_1809|M.smegmatis_MC2_155 GFLTPDD--ETVVYCRIGERSSHTWFVLTHLLGLPGVRNYDGSWTEWGNA
TH_0246|M.thermoresistible__bu GFVKPDD--KPIVYCRIGERSSHTWFVLTYLLGHPNVRNYDGSWTEWGNA
MUL_2635|M.ulcerans_Agy99 GFLDPDD--KTIVYCRIGERSSHTWFVLTHLLGKPGVRNYDGSWTEWGNT
MMAR_1253|M.marinum_M GFLDPDD--KTIVYCRIGERSSHTWFVLTHLLGKPGVRNYDGSWTEWGNT
Mb3311|M.bovis_AF2122/97 DFINPDD--QTVVYCRIGERSSHTWFVLTHLLGKADVRNYDGSWTEWGNA
Rv3283|M.tuberculosis_H37Rv DFINPDD--QTVVYCRIGERSSHTWFVLTHLLGKADVRNYDGSWTEWGNA
MLBr_00728|M.leprae_Br4923 DFITPND--KTIVYCRIGERSSHTWFVLTHLLGKPGVRNYDGSWTEWGNT
MAV_4253|M.avium_104 GFLRPDD--KTVVYCRIGERSSHTWFVLTHLLGKPGVRNYDGSWTEWGNT
MAB_3641|M.abscessus_ATCC_1997 GDVIAADG-DIVAYCRIGERSSHTWFVLTHLLGVEGVRNYDGSWTEWGNR
. : * . :.****************:*** .*************
Mflv_4780|M.gilvum_PYR-GCK VRVPVAVGSEPGDAPGEAPATAR
Mvan_1669|M.vanbaalenii_PYR-1 VRVPVAVGEEPGTA---------
MSMEG_1809|M.smegmatis_MC2_155 VRVPVAVGPDPGSAP--------
TH_0246|M.thermoresistible__bu VRVPIVVGPEPGEAPGTS-----
MUL_2635|M.ulcerans_Agy99 VRVPIVGGESPGSADGLASPR--
MMAR_1253|M.marinum_M VRVPIVGGESPGSADGLASPR--
Mb3311|M.bovis_AF2122/97 VRVPIVAGEEPGVVPVV------
Rv3283|M.tuberculosis_H37Rv VRVPIVAGEEPGVVPVV------
MLBr_00728|M.leprae_Br4923 VRVPITAGESPGAVPV-------
MAV_4253|M.avium_104 VRTPIVAGEEPGQAPAGV-----
MAB_3641|M.abscessus_ATCC_1997 VRTPIVKGDKPGAVPTQ------
**.*:. * .** .