For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DTMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGDGTQALTAVRELRPDLVLLDLMLPGMNGIDVCRVL RADSGVPIVMLTAKTDTVDVVLGLESGADDYIMKPFKPKELVARVRARLRRNDDEPAEMLSIADVDIDVP AHKATRNGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRLVNVHVQRLRAKVEKDPEN PTVVLTVRGVGYKAGPP*
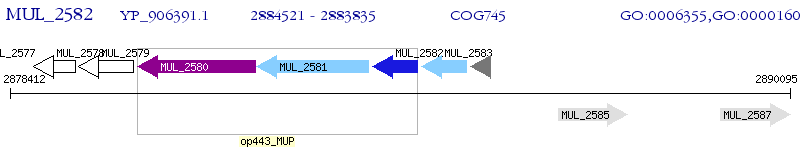
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2582 | mtrA | - | 100% (228) | two component sensory transduction transcriptional regulatory protein MtrA |
| M. ulcerans Agy99 | MUL_4560 | regX3 | 1e-56 | 48.87% (221) | two component sensory transduction protein RegX3 |
| M. ulcerans Agy99 | MUL_0248 | prrA | 6e-45 | 43.44% (221) | two component response transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3274c | mtrA | 1e-126 | 99.12% (228) | two component sensory transduction transcriptional |
| M. gilvum PYR-GCK | Mflv_4712 | - | 1e-123 | 96.93% (228) | two component transcriptional regulator |
| M. tuberculosis H37Rv | Rv3246c | mtrA | 1e-126 | 99.12% (228) | two component sensory transduction transcriptional |
| M. leprae Br4923 | MLBr_00773 | mtrA | 1e-126 | 98.68% (228) | putative two-component response regulator |
| M. abscessus ATCC 19977 | MAB_3591c | - | 1e-120 | 95.11% (225) | DNA-binding response regulator MtrA |
| M. marinum M | MMAR_1299 | mtrA | 1e-124 | 99.56% (225) | two-component sensory transduction transcriptional |
| M. avium 104 | MAV_4209 | - | 1e-125 | 98.68% (228) | DNA-binding response regulator MtrA |
| M. smegmatis MC2 155 | MSMEG_1874 | mtrA | 1e-122 | 96.05% (228) | DNA-binding response regulator MtrA |
| M. thermoresistible (build 8) | TH_4129 | - | 1e-122 | 95.18% (228) | PUTATIVE DNA-binding response regulator MtrA |
| M. vanbaalenii PYR-1 | Mvan_1752 | - | 1e-124 | 96.93% (228) | two component transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3274c|M.bovis_AF2122/97 -------MDTMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGDGTQALT
Rv3246c|M.tuberculosis_H37Rv -------MDTMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGDGTQALT
MAV_4209|M.avium_104 -------MDSMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGDGTQALT
MLBr_00773|M.leprae_Br4923 -------MDIMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGDGTQALT
MMAR_1299|M.marinum_M ----------MRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGDGTQALT
MUL_2582|M.ulcerans_Agy99 -------MDTMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGDGTQALT
MAB_3591c|M.abscessus_ATCC_199 ----------MRQRILVVDDDASLAEMLTIVLRGEGFETAVVSDGTQALT
MSMEG_1874|M.smegmatis_MC2_155 -------MDTMRQRILVVDDDPSLAEMLTIVLRGEGFDTAVIGDGSQALT
TH_4129|M.thermoresistible__bu LSRFGDNMDTMRQRILVVDDDPSLAEMLTIVLRGEGFDTAVIGDGSQALT
Mvan_1752|M.vanbaalenii_PYR-1 -------MDTMRQRILVVDDDPSLAEMLTIVLRGEGFDTAVIGDGTQALT
Mflv_4712|M.gilvum_PYR-GCK -------MDSMRQRILVVDDDPSLAEMLTIVLRGEGFDTAVIGDGTQALT
***********.***************:***:.**:****
Mb3274c|M.bovis_AF2122/97 AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTDTVDVVL
Rv3246c|M.tuberculosis_H37Rv AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTDTVDVVL
MAV_4209|M.avium_104 AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTDTVDVVL
MLBr_00773|M.leprae_Br4923 AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTDTVDVVL
MMAR_1299|M.marinum_M AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTDTVDVVL
MUL_2582|M.ulcerans_Agy99 AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTDTVDVVL
MAB_3591c|M.abscessus_ATCC_199 AVRELRPDLVLLDLMLPGMNGIDVCRVLRSDSGVPIVMLTAKTDTVDVVL
MSMEG_1874|M.smegmatis_MC2_155 AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTDTVDVVL
TH_4129|M.thermoresistible__bu AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKSDTVDVVL
Mvan_1752|M.vanbaalenii_PYR-1 AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTDTVDVVL
Mflv_4712|M.gilvum_PYR-GCK AVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTDTVDVVL
*****************************:************:*******
Mb3274c|M.bovis_AF2122/97 GLESGADDYIMKPFKPKELVARVRARLRRNDDEPAEMLSIADVEIDVPAH
Rv3246c|M.tuberculosis_H37Rv GLESGADDYIMKPFKPKELVARVRARLRRNDDEPAEMLSIADVEIDVPAH
MAV_4209|M.avium_104 GLESGADDYIMKPFKPKELVARVRARLRRNDDEPAEMLSIADVEIDVPAH
MLBr_00773|M.leprae_Br4923 GLESGADDYIMKPFKPKELVARVRARLRRNDDEPAEMLSIADVDIDVPAH
MMAR_1299|M.marinum_M GLESGADDYIMKPFKPKELVARVRARLRRNDDEPAEMLSIADVDIDVPAH
MUL_2582|M.ulcerans_Agy99 GLESGADDYIMKPFKPKELVARVRARLRRNDDEPAEMLSIADVDIDVPAH
MAB_3591c|M.abscessus_ATCC_199 GLESGADDYIMKPFKPKELVARIRARLRRNDDEPAELLNIAGIDIDVPAH
MSMEG_1874|M.smegmatis_MC2_155 GLESGADDYVMKPFKPKELVARVRARLRRNEDEPAEMLSIGDVEIDVPAH
TH_4129|M.thermoresistible__bu GLESGADDYVVKPFKPKELVARVRARLRRNEDEPAEMLSIGDIDIDVPAH
Mvan_1752|M.vanbaalenii_PYR-1 GLESGADDYVMKPFKPKELVARVRARLRRNEDEPAEMLSIADIDIDVPAH
Mflv_4712|M.gilvum_PYR-GCK GLESGADDYVMKPFKPKELVARVRARLRRNEDEPAEMLSIADVDIDVPAH
*********::***********:*******:*****:*.*..::******
Mb3274c|M.bovis_AF2122/97 KVTRNGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
Rv3246c|M.tuberculosis_H37Rv KVTRNGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
MAV_4209|M.avium_104 KVTRNGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
MLBr_00773|M.leprae_Br4923 KVTRNGEHISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
MMAR_1299|M.marinum_M KVTRNGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
MUL_2582|M.ulcerans_Agy99 KATRNGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
MAB_3591c|M.abscessus_ATCC_199 KVTREGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
MSMEG_1874|M.smegmatis_MC2_155 KVTRQGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
TH_4129|M.thermoresistible__bu KVTRNGEQIQLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
Mvan_1752|M.vanbaalenii_PYR-1 KVTRQGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
Mflv_4712|M.gilvum_PYR-GCK KVTRQGEQISLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPADTRL
*.**:**:*.****************************************
Mb3274c|M.bovis_AF2122/97 VNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGPP
Rv3246c|M.tuberculosis_H37Rv VNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGPP
MAV_4209|M.avium_104 VNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGPP
MLBr_00773|M.leprae_Br4923 VNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGPP
MMAR_1299|M.marinum_M VNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGPP
MUL_2582|M.ulcerans_Agy99 VNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGPP
MAB_3591c|M.abscessus_ATCC_199 VNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGPP
MSMEG_1874|M.smegmatis_MC2_155 VNVHVQRLRAKVEKDPENPQVVLTVRGVGYKAGPP
TH_4129|M.thermoresistible__bu VNVHVQRLRAKVEKDPENPQVVLTVRGVGYKAGPP
Mvan_1752|M.vanbaalenii_PYR-1 VNVHVQRLRAKVEKDPENPQVVLTVRGVGYKAGPP
Mflv_4712|M.gilvum_PYR-GCK VNVHVQRLRAKVEKDPENPQVVLTVRGVGYKAGPP
******************* ***************