For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRLEQMYQDVILDHYKHPQHRGLREPFATQVSHVNPVCGDEVTLRVTLSNDGATIADVSYDGQGCSISQA ATSVLTQQVIGQTVPEALETVDAFSQMVSSRGTVQGDEDVLGDGVAFAGVAKYPARVKCALLGWMAFKDA LAQAITANALAEGGGHVEEVSDE
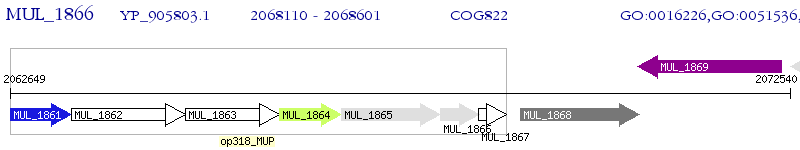
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1866 | - | - | 100% (163) | nitrogen fixation related protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1500 | - | 6e-71 | 80.37% (163) | nitrogen fixation related protein |
| M. gilvum PYR-GCK | Mflv_3673 | - | 6e-65 | 78.47% (144) | NifU family SUF system FeS assembly protein |
| M. tuberculosis H37Rv | Rv1465 | - | 6e-71 | 80.37% (163) | nitrogen fixation related protein |
| M. leprae Br4923 | MLBr_00597 | - | 6e-68 | 73.62% (163) | hypothetical protein MLBr_00597 |
| M. abscessus ATCC 19977 | MAB_2745c | - | 8e-64 | 78.62% (145) | SUF system FeS assembly protein |
| M. marinum M | MMAR_2270 | - | 9e-90 | 99.39% (163) | nitrogen fixation related protein |
| M. avium 104 | MAV_3314 | - | 2e-70 | 79.49% (156) | SUF system FeS assembly protein, NifU family protein |
| M. smegmatis MC2 155 | MSMEG_3126 | - | 7e-68 | 77.85% (158) | SUF system FeS assembly protein, NifU family protein |
| M. thermoresistible (build 8) | TH_1612 | - | 1e-57 | 73.47% (147) | POSSIBLE NITROGEN FIXATION RELATED PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_2737 | - | 2e-66 | 74.85% (163) | NifU family SUF system FeS assembly protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_1866|M.ulcerans_Agy99 --MRLEQMYQDVILDHYKHPQHRGLREPFATQVSHVNPVCGDEVTLRVTL
MMAR_2270|M.marinum_M --MRLEQMYQDVILDHYKHPQHRGLREPFATQVSHVNPVCGDEVTLRVTL
Mflv_3673|M.gilvum_PYR-GCK --MRLEQIYQEVILDHYKHPHHRGLREPFAAEVHHVNPTCGDEVTLRVAV
Mvan_2737|M.vanbaalenii_PYR-1 --MRLEQIYQEVILDHYKHPHHRGLREPFAAEVHHVNPTCGDEVTLRVTL
MSMEG_3126|M.smegmatis_MC2_155 --MRLEQMYQEVILDHYKHPHHRGLREPFGAEVHHVNPTCGDEVTLRVTL
MAB_2745c|M.abscessus_ATCC_199 --MRLEQMYQEVILDHYKHPHHRGLREPFAAEVHHVNPTCGDEVTLRVFV
TH_1612|M.thermoresistible__bu -------MYQDVILEHYKHPLNRGLREPFDAESYQVNPTCGDEVRLRVTL
Mb1500|M.bovis_AF2122/97 MTLRLEQIYQDVILDHYKHPQHRGLREPFGAQVYHVNPICGDEVTLRVAL
Rv1465|M.tuberculosis_H37Rv MTLRLEQIYQDVILDHYKHPQHRGLREPFGAQVYHVNPICGDEVTLRVAL
MLBr_00597|M.leprae_Br4923 MILRLEQIYQEVILDHYKHPQHRGLREPFCAQVYHVNPICGDEITLRVAL
MAV_3314|M.avium_104 --MRLEQMYQDVILDHYKHPQHRGLREPFGAEVFHVNPVCGDEVTLRVTL
:**:***:***** :******* :: :*** ****: *** :
MUL_1866|M.ulcerans_Agy99 SNDGATIADVSYDGQGCSISQAATSVLTQQVIGQTVPEALETVDAFSQMV
MMAR_2270|M.marinum_M SNDGATIADVSYDGQGCSISQAATSVLTQQVIGQTVPEALETVDAFSQMV
Mflv_3673|M.gilvum_PYR-GCK SDDGETVTDISYDGQGCSISQASTSVLTDQVIGQTVGDALKTVAAFTEMV
Mvan_2737|M.vanbaalenii_PYR-1 SEDGETVTDISYDGQGCSISQAATSVLTDQVIGQSVGDALKTVAAFSEMV
MSMEG_3126|M.smegmatis_MC2_155 SDDGESVEDVSYDGQGCSISQAATSVLTDQVIGQSVGDALKTVDAFTEMV
MAB_2745c|M.abscessus_ATCC_199 QDN--KIADVSYDGQGCSISQAATSVLTDQVIGLTVDEALKTVASFNEMI
TH_1612|M.thermoresistible__bu GDG-DVVRDVSYEGQGCSISQAATSVLTDLVIGQKVEDALRTVAAFTEMV
Mb1500|M.bovis_AF2122/97 SEDGTRVTDVSYDGQGCSISQAATSVLTEQVIGQRVPRALNIVDAFTEMV
Rv1465|M.tuberculosis_H37Rv SEDGTRVTDVSYDGQGCSISQAATSVLTEQVIGQRVPRALNIVDAFTEMV
MLBr_00597|M.leprae_Br4923 SDNGASVADISYEGQGCSISQASISVLAQQVIGQSVPDALNIISAFTEMV
MAV_3314|M.avium_104 SDDGETVADVSYDGQGCSISQAATSVLTEQVIGRSVEEALDTIGAFAEMV
:. : *:**:*********: ***:: *** * ** : :* :*:
MUL_1866|M.ulcerans_Agy99 SSRGTVQGDEDVLGDGVAFAGVAKYPARVKCALLGWMAFKDALAQAITAN
MMAR_2270|M.marinum_M SSRGTVQGDEDVLGDGVAFAGVAKYPARVKCALLGWMAFKDALAQAITAN
Mflv_3673|M.gilvum_PYR-GCK SSRGKIDGDEDVLGDGIAFAGVSKYPARVKCALLGWTAFKAALAEA----
Mvan_2737|M.vanbaalenii_PYR-1 SSRGKVDGDEDVLGDGIAFAGVSKYPARVKCALLGWTAFKAALAQA----
MSMEG_3126|M.smegmatis_MC2_155 SSRGTVEGDEDVIGDGIAFAGVAKYPARVKCALLGWMAFKAAVAEARPLD
MAB_2745c|M.abscessus_ATCC_199 SSRGTIDGDEDVIGDGIAFAGVSRYPARVKCALLGWMAFKDALVQA----
TH_1612|M.thermoresistible__bu TSRGKVDGDEDVIGDGIAFAGVAKYPARVKCALLGWMAFKDAVAQA----
Mb1500|M.bovis_AF2122/97 SSRGTVPGDEDVLGDGVAFAGVAKYPARVKCALLGWMAFKDALAQAS---
Rv1465|M.tuberculosis_H37Rv SSRGTVPGDEDVLGDGVAFAGVAKYPARVKCALLGWMAFKDALAQAS---
MLBr_00597|M.leprae_Br4923 SSRGTIEGDEDVLGDGVAFAGVAKYPARVKCALLGWMACKDALIHANSA-
MAV_3314|M.avium_104 SSRGAVEGDEDVLGDGIAFAGVAKYPARVKCALLGWMACKDALAQAY---
:*** : *****:***:*****::************ * * *: .*
MUL_1866|M.ulcerans_Agy99 ALAEGGGHVEEVSDE---
MMAR_2270|M.marinum_M ALAEGGEHVEEVSDE---
Mflv_3673|M.gilvum_PYR-GCK --HG--PENSEGEST---
Mvan_2737|M.vanbaalenii_PYR-1 --HG--PEISE-EN----
MSMEG_3126|M.smegmatis_MC2_155 ETHATLAEAGEREDNR--
MAB_2745c|M.abscessus_ATCC_199 ------IEHHEVKR----
TH_1612|M.thermoresistible__bu ------ARTQDLEEAR--
Mb1500|M.bovis_AF2122/97 ---EAFEEVTDERNQRTG
Rv1465|M.tuberculosis_H37Rv ---EAFEEVTDERNQRTG
MLBr_00597|M.leprae_Br4923 --DKVLEEVMDEQNHRVG
MAV_3314|M.avium_104 ----SRREVTDERNHRAG
. :