For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTPRHLLAAGDLSRDDAIAILDDADRFAQALVGREVKKLPTLRGRTVVTMFYENSTRTRVSFEVAGKWMS ADLINVSASGSSVSKGESLRDTALTLRAAGADALIIRHPASGAARLFADWTAGQSDGGPSVINAGDGTHE HPTQALLDALTIRQRLGGIEGRRVVIVGDILHSRVARSNVTLLHTLGAEVVLVAPPTLLPVGVADWPVTV SHDLDAELPAADAVLMLRVQAERMNGGFFPSVREYSTLYGLSDRRQAMLGGHAVVLHPGPMLRGMEIASS VADSSQSAVLQQVSNGVHIRMAVLFHVLVGLESAGEEGAA
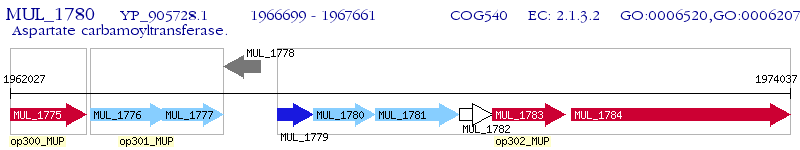
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1780 | pyrB | - | 100% (320) | aspartate carbamoyltransferase catalytic subunit |
| M. ulcerans Agy99 | MUL_1648 | argF | 3e-16 | 29.11% (316) | ornithine carbamoyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1415 | pyrB | 1e-158 | 88.44% (320) | aspartate carbamoyltransferase catalytic subunit |
| M. gilvum PYR-GCK | Mflv_3747 | pyrB | 1e-149 | 86.08% (309) | aspartate carbamoyltransferase catalytic subunit |
| M. tuberculosis H37Rv | Rv1380 | pyrB | 1e-158 | 88.44% (320) | aspartate carbamoyltransferase catalytic subunit |
| M. leprae Br4923 | MLBr_00532 | pyrB | 1e-155 | 87.42% (318) | aspartate carbamoyltransferase catalytic subunit |
| M. abscessus ATCC 19977 | MAB_2831c | pyrB | 1e-147 | 83.39% (313) | aspartate carbamoyltransferase catalytic subunit |
| M. marinum M | MMAR_2195 | pyrB | 1e-177 | 99.06% (320) | aspartate carbamoyltransferase PyrB |
| M. avium 104 | MAV_3393 | pyrB | 1e-159 | 91.99% (312) | aspartate carbamoyltransferase catalytic subunit |
| M. smegmatis MC2 155 | MSMEG_3043 | pyrB | 1e-150 | 84.29% (312) | aspartate carbamoyltransferase catalytic subunit |
| M. thermoresistible (build 8) | TH_1351 | pyrB | 1e-155 | 87.82% (312) | PROBABLE ASPARTATE CARBAMOYLTRANSFERASE PYRB (ATCase) |
| M. vanbaalenii PYR-1 | Mvan_2660 | pyrB | 1e-149 | 85.76% (309) | aspartate carbamoyltransferase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3747|M.gilvum_PYR-GCK --MKHLLTAADLSRDDATAILDNADRFREALVGREVKKLPTLRGRTIITM
Mvan_2660|M.vanbaalenii_PYR-1 --MKHLLTAADLSREDATAILDNADRFREALVGREVKKLPTLRGRTIITM
MSMEG_3043|M.smegmatis_MC2_155 MTKRHLLSAGDLTRDDATAILDDADRFREALLGREVKKLPTLRGRTIITM
MUL_1780|M.ulcerans_Agy99 MTPRHLLAAGDLSRDDAIAILDDADRFAQALVGREVKKLPTLRGRTVVTM
MMAR_2195|M.marinum_M MTPRHLLAAGDLSRDDAIAILDDADRFAQALVGREVKKLPTLRGRTVVTM
Mb1415|M.bovis_AF2122/97 MTPRHLLTAADLSRDDATAILDDADRFAQALVGRDIKKLPTLRGRTVVTM
Rv1380|M.tuberculosis_H37Rv MTPRHLLTAADLSRDDATAILDDADRFAQALVGRDIKKLPTLRGRTVVTM
MLBr_00532|M.leprae_Br4923 MMRRHLLNAGDLSRDDAIAIFDDADRFAHALVGREIKKLPTLRGRTVITM
MAV_3393|M.avium_104 -MTRHLLAAGDLSRDDATAILDDADRFAQALVGREIKKLPTLRGRTVVTM
TH_1351|M.thermoresistible__bu MTARHLLSAADLSRDEAVAILDDADRFRQALLGRDVKKLPTLRGRTIITM
MAB_2831c|M.abscessus_ATCC_199 --MRHLLSAADLTRDEATAILDDADRFLQALAGREVKKLPTLRGRTIITM
:*** *.**:*::* **:*:**** .** **::**********::**
Mflv_3747|M.gilvum_PYR-GCK FYENSTRTRVSFEVAGKWMSADVINVSASGSSVAKGESLRDTALTLRAAG
Mvan_2660|M.vanbaalenii_PYR-1 FYENSTRTRVSFEVAGKWMSADVINVSSSGSSVAKGESLRDTALTLRAAG
MSMEG_3043|M.smegmatis_MC2_155 FYENSTRTRVSFEVAGKWMSADVINVSASGSSVAKGESLRDTALTLRAAG
MUL_1780|M.ulcerans_Agy99 FYENSTRTRVSFEVAGKWMSADLINVSASGSSVSKGESLRDTALTLRAAG
MMAR_2195|M.marinum_M FYENSTRTRVSFEVAGKWMSADVINVSASGSSVSKGESLRDTALTLRAAG
Mb1415|M.bovis_AF2122/97 FYENSTRTRVSFEVAGKWMSADVINVSAAGSSVGKGESLRDTALTLRAAG
Rv1380|M.tuberculosis_H37Rv FYENSTRTRVSFEVAGKWMSADVINVSAAGSSVGKGESLRDTALTLRAAG
MLBr_00532|M.leprae_Br4923 FYENSTRTRVSFEVAGKWMSANVINVSPAGSSVGKGESLRDTALTLRAAG
MAV_3393|M.avium_104 FYENSTRTRVSFEVAGKWMSADVINVSAAGSSVGKGESLRDTALTLRAAG
TH_1351|M.thermoresistible__bu FYENSTRTRVSFEVAGKWMSADVINVSATGSSVGKGESLRDTALTLRAAG
MAB_2831c|M.abscessus_ATCC_199 FYENSTRTRVSFEVAGKWMSADVINVSASGSSVAKGESLRDTALTLRAIG
*********************::****.:****.************** *
Mflv_3747|M.gilvum_PYR-GCK ADALIIRHPASGAPQQLAAWTL---DEHGG-PSVINAGDGTHEHPTQALL
Mvan_2660|M.vanbaalenii_PYR-1 ADALIIRHPASGAAQQLAAWTL---DEHGG-PSIINAGDGTHEHPTQALL
MSMEG_3043|M.smegmatis_MC2_155 ADALIIRHPASGAAQQLAEWTV---EEDGGGPSVINAGDGTHEHPTQALL
MUL_1780|M.ulcerans_Agy99 ADALIIRHPASGAARLFADWTA--GQS-DGGPSVINAGDGTHEHPTQALL
MMAR_2195|M.marinum_M ADALIIRHPASGAARLLADWTA--GQS-DGGPSVINAGDGTHEHPTQALL
Mb1415|M.bovis_AF2122/97 ADALIIRHPASGAAHLLAQWTG--AHN--DGPAVINAGDGTHEHPTQALL
Rv1380|M.tuberculosis_H37Rv ADALIIRHPASGAAHLLAQWTG--AHN--DGPAVINAGDGTHEHPTQALL
MLBr_00532|M.leprae_Br4923 ADALIIRHPASGAAHLLADWTA--AETGEDGPAVINAGDGTHEHPTQALL
MAV_3393|M.avium_104 ADALIIRHPASGAAHLLAEWTCGNGDSSDAGPSVINAGDGTHEHPTQALL
TH_1351|M.thermoresistible__bu ADALVIRHPASGAAQQLAHWTAD-GDG--GGPSVINAGDGTHEHPTQALL
MAB_2831c|M.abscessus_ATCC_199 ADALIVRHPASGVAAQLAQWTA----EGEGGPAVVNAGDGTHEHPTQALL
****::******.. :* ** *:::***************
Mflv_3747|M.gilvum_PYR-GCK DALTIRQRLGSVEGKRVVIVGDVLHSRVARSNVALLHTLGAEVVLVAPPT
Mvan_2660|M.vanbaalenii_PYR-1 DALTIRQRLGSVEGKRVVIVGDVLHSRVARSNVALLHTLGAEVVLVAPPT
MSMEG_3043|M.smegmatis_MC2_155 DALTIRQRLGSVEGKRVVIVGDVLHSRVARSNVTLLHTLGAEVVLVSPPT
MUL_1780|M.ulcerans_Agy99 DALTIRQRLGGIEGRRVVIVGDILHSRVARSNVTLLHTLGAEVVLVAPPT
MMAR_2195|M.marinum_M DALTIRQRLGGIEGRRVVIVGDILHSRVARSNVMLLHTLGAEVVLVAPPT
Mb1415|M.bovis_AF2122/97 DALTIRQRLGGIEGRRIVIVGDILHSRVARSNVMLLDTLGAEVVLVAPPT
Rv1380|M.tuberculosis_H37Rv DALTIRQRLGGIEGRRIVIVGDILHSRVARSNVMLLDTLGAEVVLVAPPT
MLBr_00532|M.leprae_Br4923 DALTIRQRLGSIEGRRILIVGDILHSRVARSNVVLLDTLGAEVVLVAPRT
MAV_3393|M.avium_104 DALTIRQRLGGIEGRRVVIVGDVLHSRVARSNVMLLSTLGAEVVLVAPPT
TH_1351|M.thermoresistible__bu DALTIRQRLGGIEGRRVVIVGDILHSRVARSNVSLLHTLGAEVVLVAPPT
MAB_2831c|M.abscessus_ATCC_199 DALTIRQRLGSLEGRRVVIVGDILHSRVARSNALLLNTFGAEVVLVAPPT
**********.:**:*::****:*********. ** *:*******:* *
Mflv_3747|M.gilvum_PYR-GCK LLPVGVTEWPVTVSHELDAELPLADAVLMLRVQAERMNGGFFPSSREYSV
Mvan_2660|M.vanbaalenii_PYR-1 LLPVGVADWPVTVSQDIDAELPLADAVLMLRVQAERMNGGFFPSSREYSV
MSMEG_3043|M.smegmatis_MC2_155 LLPRGVENWPVTVSYDLDAELPAADAVLMLRVQAERMNGSFFPSAREYSV
MUL_1780|M.ulcerans_Agy99 LLPVGVADWPVTVSHDLDAELPAADAVLMLRVQAERMNGGFFPSVREYST
MMAR_2195|M.marinum_M LLPVGVADWPVTVSHDLDAELPAADAVLMLRVQAERMNGGFFPSVREYST
Mb1415|M.bovis_AF2122/97 LLPVGVTGWPATVSHDFDAELPAADAVLMLRVQAERMNGGFFPSVREYSV
Rv1380|M.tuberculosis_H37Rv LLPVGVTGWPATVSHDFDAELPAADAVLMLRVQAERMNGGFFPSVREYSV
MLBr_00532|M.leprae_Br4923 LLPIGVDGWPATVSCDFDAELPAADAVLMLRVQAERMNGGFFPSVREYSC
MAV_3393|M.avium_104 LLPVGVEGWPVTVSNDFDAELPAADAVLMLRVQAERMNGGFFPSVREYSS
TH_1351|M.thermoresistible__bu LLPVGVTDWPATVSHDLDAELPGADAVLMLRVQAERMNGGFFPSAREYSV
MAB_2831c|M.abscessus_ATCC_199 LLPVGVGTWPVTVTHDLDAELPGADAVLMLRVQAERMTGGFFPSAREYSV
*** ** **.**: ::***** **************.*.**** ****
Mflv_3747|M.gilvum_PYR-GCK RYGLSEKRQSTLADHAVVLHPGPMLRGMEIAYSVADSSQSAVLQQVSNGV
Mvan_2660|M.vanbaalenii_PYR-1 RYGLSEKRQSRLAEHAVVLHPGPMLRGMEIAYSVADSSQSAVLQQVSNGV
MSMEG_3043|M.smegmatis_MC2_155 RYGLSEKRQAMLPDRAIVLHPGPMVRGMEISFPVADSPQSAVLQQVSNGV
MUL_1780|M.ulcerans_Agy99 LYGLSDRRQAMLGGHAVVLHPGPMLRGMEIASSVADSSQSAVLQQVSNGV
MMAR_2195|M.marinum_M LYGLSDRRQAMLGGHAVVLHPGPMLRGMEIASSVADSSQSAVLQQVSNGV
Mb1415|M.bovis_AF2122/97 RYGLTERRQAMLPGHAVVLHPGPMVRGMEITSSVADSSQSAVLQQVSNGV
Rv1380|M.tuberculosis_H37Rv RYGLTERRQAMLPGHAVVLHPGPMVRGMEITSSVADSSQSAVLQQVSNGV
MLBr_00532|M.leprae_Br4923 LYGLTERRQALLPGHAVVLHPGPMLRGMEIASSVADSSQSAVLQQVSNGV
MAV_3393|M.avium_104 LYGLSDRRQAMLPGHAVVLHPGPMLRGMEIASSVADSSQSAVLQQVSNGV
TH_1351|M.thermoresistible__bu LYGLSEQRQAMLSGDAVVLHPGPMLRGMEIASTVADSVQSAVLQQVSNGV
MAB_2831c|M.abscessus_ATCC_199 LYGMSEARQRLLPEHAVVLHPGPMLRGMEIASAVADSSQSGVLQQVSNGV
**::: ** * *:*******:*****: .**** **.*********
Mflv_3747|M.gilvum_PYR-GCK HVRMAVLFHLLVGSEEEAISA--
Mvan_2660|M.vanbaalenii_PYR-1 HVRMAVLFHLLVGSEQEAISA--
MSMEG_3043|M.smegmatis_MC2_155 HVRMAVLFHLLVGADREAINV--
MUL_1780|M.ulcerans_Agy99 HIRMAVLFHVLVGLESAGEEGAA
MMAR_2195|M.marinum_M HIRMAVLFHVLVGLESAGEEGAA
Mb1415|M.bovis_AF2122/97 QVRMAVLFHVLVGAQDAGKEGAA
Rv1380|M.tuberculosis_H37Rv QVRMAVLFHVLVGAQDAGKEGAA
MLBr_00532|M.leprae_Br4923 HVRMAVLFHVLVGTQSAEEEGAA
MAV_3393|M.avium_104 HVRMAVLFHVLVGAEEIAA----
TH_1351|M.thermoresistible__bu HVRMAVLFHLLVGADEQVSV---
MAB_2831c|M.abscessus_ATCC_199 HIRMAVLFHLLVGSSDSPEAVPA
::*******:*** .