For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSDNKEPVTVQVAVDPPYPVVIGTGLLTELEELLAGRHKVAILHQPVLTETAEAIRNHLADKGIDAHRIE IPDAEAGKELPVVVFIWEVLGRIGIGRKDALVSLGGGAATDVAGFAAATWLRGVSIVHVPTTLLGMVDAA VGGKTGINTDAGKNLVGAFHQPLGVLADLATLQTLPQNEIVCGMAEVVKAGFIADPVILDLIEADPQAAL DPSGDVMPELIRRSIAVKAEVVAADEKESELREILNYGHTLAHAIERRERYQWQHGAAVSVGLVFAAELA RLSGRLDDAMALRHRTILSALGLPVSYDADALPQLLEFMTGDKKTRAGVLRFVVLDGLAKPGRMAGPDPG LLVTAYAGICAP
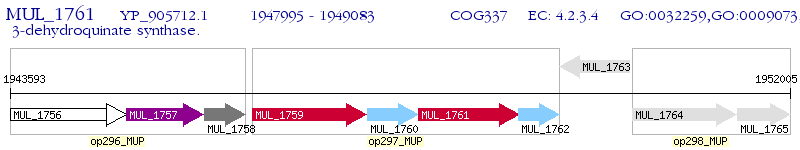
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1761 | aroB | - | 100% (362) | 3-dehydroquinate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2567c | aroB | 0.0 | 88.67% (362) | 3-dehydroquinate synthase |
| M. gilvum PYR-GCK | Mflv_3756 | aroB | 1e-170 | 83.05% (354) | 3-dehydroquinate synthase |
| M. tuberculosis H37Rv | Rv2538c | aroB | 0.0 | 88.67% (362) | 3-dehydroquinate synthase |
| M. leprae Br4923 | MLBr_00518 | aroB | 1e-177 | 85.32% (361) | 3-dehydroquinate synthase |
| M. abscessus ATCC 19977 | MAB_2841c | aroB | 1e-156 | 77.97% (354) | 3-dehydroquinate synthase |
| M. marinum M | MMAR_2177 | aroB | 0.0 | 98.62% (362) | 3-dehydroquinate synthase AroB |
| M. avium 104 | MAV_3415 | aroB | 1e-174 | 86.36% (352) | 3-dehydroquinate synthase |
| M. smegmatis MC2 155 | MSMEG_3033 | aroB | 1e-169 | 84.00% (350) | 3-dehydroquinate synthase |
| M. thermoresistible (build 8) | TH_0108 | aroB | 1e-175 | 84.55% (356) | 3-DEHYDROQUINATE SYNTHASE AROB |
| M. vanbaalenii PYR-1 | Mvan_2647 | aroB | 1e-173 | 84.46% (354) | 3-dehydroquinate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3756|M.gilvum_PYR-GCK ---MTEPVTVDVRTDPPYPVIIGRGLLGDLGRVLDGRH---KVAILHQPT
Mvan_2647|M.vanbaalenii_PYR-1 ---MTQPVTVEVRTDPPYPVIIGTGLLDDLARVLDGRH---KVAILHQPT
MSMEG_3033|M.smegmatis_MC2_155 -------MIVEVKVDRPYPVVIGTGLLEDLGRTLEGRH---KVAVLHQPV
TH_0108|M.thermoresistible__bu ---MTEPVTVDVQVDPPYPVIIGTGLLGELDRLLGGRH---KVAVLHQPV
MUL_1761|M.ulcerans_Agy99 MSDNKEPVTVQVAVDPPYPVVIGTGLLTELEELLAGRH---KVAILHQPV
MMAR_2177|M.marinum_M MSDNKEPVTVQVAVDPPYPVVIGTGLLTELEELLAGRH---KVAILHQPV
Mb2567c|M.bovis_AF2122/97 MTDIGAPVTVQVAVDPPYPVVIGTGLLDELEDLLADRH---KVAVVHQPG
Rv2538c|M.tuberculosis_H37Rv MTDIGAPVTVQVAVDPPYPVVIGTGLLDELEDLLADRH---KVAVVHQPG
MLBr_00518|M.leprae_Br4923 MTNIDAPVTVQVAVDPPYPVVIGTGLSDQLDELLANRH---RVAILHQPV
MAV_3415|M.avium_104 -------MTVQVAVDPPYPVIIGTGLLGELDELLGARH---KVAVVHQPV
MAB_2841c|M.abscessus_ATCC_199 -MTSANPTTITVEVNPPYPVIVGTGLLGELVDNLKGSQGLAKVAILHQPT
: * .: ****::* ** :* * : :**::***
Mflv_3756|M.gilvum_PYR-GCK LTQTAEAIRTHLSEKGIDAHRIEIPDAEGGKELPVVGFIWQVLGRIGVGR
Mvan_2647|M.vanbaalenii_PYR-1 LTQTAEVIRNHLADKGIDAHRIEIPDAEGGKELPVVGFIWEVLGRIGLGR
MSMEG_3033|M.smegmatis_MC2_155 LNQTAEAIRSHLADKGIDAHRIELPDAEAGKELPVVGFIWEVLGRIGIGR
TH_0108|M.thermoresistible__bu LAQTAEAIRSRLADSGVDAHRIEIPDAEAGKDLPVVGFVWEVLGRIGLGR
MUL_1761|M.ulcerans_Agy99 LTETAEAIRNHLADKGIDAHRIEIPDAEAGKELPVVVFIWEVLGRIGIGR
MMAR_2177|M.marinum_M LTETAEAIRNHLADKGIDAHRIEIPDAEAGKELPVVGFIWEVLGRIGIGR
Mb2567c|M.bovis_AF2122/97 LAETAEEIRKRLAGKGVDAHRIEIPDAEAGKDLPVVGFIWEVLGRIGIGR
Rv2538c|M.tuberculosis_H37Rv LAETAEEIRKRLAGKGVDAHRIEIPDAEAGKDLPVVGFIWEVLGRIGIGR
MLBr_00518|M.leprae_Br4923 LTQTAEAIRSHLAGKGVDAHRIEIPDAEAGKDLSVMDFIWEVLGRIGIGR
MAV_3415|M.avium_104 LAETAEVIRKRLADKGVDAHRIEIPDAEDGKDLPVVGFLWEVLGRIGIGR
MAB_2841c|M.abscessus_ATCC_199 LAVTAEEIRKTLADSGIDAHRVEIPDAEAGKDLAVLGFIWEVLGRIGIGR
* *** **. *: .*:****:*:**** **:*.*: *:*:******:**
Mflv_3756|M.gilvum_PYR-GCK KDAIVSLGGGAATDVAGFAAATWLRGIDIVHVPTTLLGMVDAAVGGKTGI
Mvan_2647|M.vanbaalenii_PYR-1 KDAIVSLGGGAATDVAGFAAATWLRGVDIVHVPTTLLGMVDAAVGGKTGI
MSMEG_3033|M.smegmatis_MC2_155 KDALVSLGGGAATDVAGFAAATWLRGVDIVHVPTTLLGMVDAAVGGKTGI
TH_0108|M.thermoresistible__bu KDAIVSLGGGAATDVAGFVAATWLRGVDIVHIPTTLLGMVDAAVGGKTGI
MUL_1761|M.ulcerans_Agy99 KDALVSLGGGAATDVAGFAAATWLRGVSIVHVPTTLLGMVDAAVGGKTGI
MMAR_2177|M.marinum_M KDALVSLGGGAATDVAGFAAATWLRGVSIVHVPTTLLGMVDAAVGGKTGI
Mb2567c|M.bovis_AF2122/97 KDALVSLGGGAATDVAGFAAATWLRGVSIVHLPTTLLGMVDAAVGGKTGI
Rv2538c|M.tuberculosis_H37Rv KDALVSLGGGAATDVAGFAAATWLRGVSIVHLPTTLLGMVDAAVGGKTGI
MLBr_00518|M.leprae_Br4923 KDALVSFGGGAATDVAGFAAATWLRGVSIVHVPTTLLGMVDAAVGGKTGI
MAV_3415|M.avium_104 KDALVSLGGGAATDVAGFAAATWLRGVDIVHVPTTLLGMVDAAVGGKTGI
MAB_2841c|M.abscessus_ATCC_199 KDAVVSLGGGAATDTAGFAAATWLRGIKVVHVPTTLLAMVDAAVGGKTGI
***:**:*******.***.*******:.:**:*****.************
Mflv_3756|M.gilvum_PYR-GCK NTDAGKNLVGAFHQPAAVLIDLATLESLPRNEIVAGMAEIVKAGFIADPV
Mvan_2647|M.vanbaalenii_PYR-1 NTDAGKNLVGAFHQPAAVLIDLATLESLPRNEIVAGMAEIVKAGFIADPV
MSMEG_3033|M.smegmatis_MC2_155 NTDAGKNLVGAFHQPAAVLVDLATLETLPRNEIVAGMAEIVKAGFIADPV
TH_0108|M.thermoresistible__bu NTDAGKNLVGAFHQPIGVLIDLATMETLPRNEIVAGMAEVVKAGFIADPK
MUL_1761|M.ulcerans_Agy99 NTDAGKNLVGAFHQPLGVLADLATLQTLPQNEIVCGMAEVVKAGFIADPV
MMAR_2177|M.marinum_M NTDAGKNLVGAFHQPLAVLADLATLQTLPQNEIVCGMAEVVKAGFIADPV
Mb2567c|M.bovis_AF2122/97 NTDAGKNLVGAFHQPLAVLVDLATLQTLPRDEMICGMAEVVKAGFIADPV
Rv2538c|M.tuberculosis_H37Rv NTDAGKNLVGAFHQPLAVLVDLATLQTLPRDEMICGMAEVVKAGFIADPV
MLBr_00518|M.leprae_Br4923 NTEAGKNLVGAFHQPLAVLADLATLETLPRKEIASGMAEVVKAGFIADPI
MAV_3415|M.avium_104 NTDAGKNLVGAFHQPLAVLVDLATLHTLPPNELVAGMAEVVKAGFIADPA
MAB_2841c|M.abscessus_ATCC_199 NTEAGKNLVGAFHQPGAVLVDLATLESLPRNEIIAGMAEVVKAGFIADPV
**:************ .** ****:.:** .*: .****:*********
Mflv_3756|M.gilvum_PYR-GCK ILDMIEADPEAALDPSGAVLPELIRRAVVVKAEVVAADEKESQLREILNY
Mvan_2647|M.vanbaalenii_PYR-1 ILDMIEADPQAALDPTGQVLPELIRRAVVVKAEVVAADEKESQLREILNY
MSMEG_3033|M.smegmatis_MC2_155 ILDLIEADPEAAIDPTGSVLPELIRRAVAVKADVVAADEKESALREILNY
TH_0108|M.thermoresistible__bu ILDLIEANPQAALDPAGDVLPELIRRAVAVKAEVVAADEKESQLREILNY
MUL_1761|M.ulcerans_Agy99 ILDLIEADPQAALDPSGDVMPELIRRSIAVKAEVVAADEKESELREILNY
MMAR_2177|M.marinum_M ILDLIEADPQAALDPSGDVMPELIRRSIAVKAEVVAADEKESELREILNY
Mb2567c|M.bovis_AF2122/97 ILDLIEADPQAALDPAGDVLPELIRRAITVKAEVVAADEKESELREILNY
Rv2538c|M.tuberculosis_H37Rv ILDLIEADPQAALDPAGDVLPELIRRAITVKAEVVAADEKESELREILNY
MLBr_00518|M.leprae_Br4923 ILDLIEADPQASLDPMGGVLPELIRRAVTVKAGVVSADEKESELREILNY
MAV_3415|M.avium_104 ILDLIEADPRAAVDPSGEVLAELVQRAIAVKADVVAADEKESELREILNY
MAB_2841c|M.abscessus_ATCC_199 ILDLIEADPETAIDPSKPVLRELVERSIAVKATVVAADEKEAELREILNY
***:***:*.:::** *: **:.*::.*** **:*****: *******
Mflv_3756|M.gilvum_PYR-GCK GHTLAHAIERRERYQWRHGAAVSVGLVFAAELGRLAGRLDDDTTERHRAI
Mvan_2647|M.vanbaalenii_PYR-1 GHTLAHAIERRERYQWRHGAAVSVGLVFAAELGRLAGRLDDATAERHRAI
MSMEG_3033|M.smegmatis_MC2_155 GHTLAHAIERRERYKWRHGAAVSVGLVFAAELGRLAGGLGDQTADRHRSV
TH_0108|M.thermoresistible__bu GHTLAHAIERRERYQWRHGAAVSVGLVYAAELGRLAGRLDDATADRHRSV
MUL_1761|M.ulcerans_Agy99 GHTLAHAIERRERYQWQHGAAVSVGLVFAAELARLSGRLDDAMALRHRTI
MMAR_2177|M.marinum_M GHTLAHAIERRERYQWRHGAAVSVGLVFAAELARLSGRLDDATALRHRTI
Mb2567c|M.bovis_AF2122/97 GHTLGHAIERRERYRWRHGAAVSVGLVFAAELARLAGRLDDATAQRHRTI
Rv2538c|M.tuberculosis_H37Rv GHTLGHAIERRERYRWRHGAAVSVGLVFAAELARLAGRLDDATAQRHRTI
MLBr_00518|M.leprae_Br4923 GHTLAHAIERRERYEWRHGAAVSVGLVFAAELARVAGRLDDATAQRHHTI
MAV_3415|M.avium_104 GHTLGHAIERRERYRWRHGAAVSVGLVFAAELARLAGRLDDATARRHRTI
MAB_2841c|M.abscessus_ATCC_199 GHTLGHAIERRERYRWRHGAAVSVGLVFAAELGRLAGRLDDETADRHARV
****.*********.*:**********:****.*::* *.* : ** :
Mflv_3756|M.gilvum_PYR-GCK LTSLGLPVTYDADALPQLMESMLGDKKTRAGVLRFVVLDGLAKPGRLEGP
Mvan_2647|M.vanbaalenii_PYR-1 LISLGLPVTYDADALPQLMEAMLGDKKTRAGVLRFVVLDGLGKPGRLEGP
MSMEG_3033|M.smegmatis_MC2_155 LTALGLPITYDADALPQLLEYMAGDKKNRSGVLRFVVLDGLAKPGRLEGP
TH_0108|M.thermoresistible__bu LTALGLPVSYDADALPELLEFMRGDKKTRSGVLRFVVLDGLAKPGRLEGP
MUL_1761|M.ulcerans_Agy99 LSALGLPVSYDADALPQLLEFMTGDKKTRAGVLRFVVLDGLAKPGRMAGP
MMAR_2177|M.marinum_M LSALGLPVSYDADALPQLLEFMTGDKKTRAGVLRFVVLDGLAKPGRMVGP
Mb2567c|M.bovis_AF2122/97 LSSLGLPVSYDPDALPQLLEIMAGDKKTRAGVLRFVVLDGLAKPGRMVGP
Rv2538c|M.tuberculosis_H37Rv LSSLGLPVSYDPDALPQLLEIMAGDKKTRAGVLRFVVLDGLAKPGRMVGP
MLBr_00518|M.leprae_Br4923 LTSLGLPVSYDADALPQLLEYMAGDKKTRAGVLRFVILDGLAKPGRLVGP
MAV_3415|M.avium_104 LSALGLPVSYDADALPQLLEYMAGDKKSRAGVLRFVVLDGLAKPGRLAGP
MAB_2841c|M.abscessus_ATCC_199 LSALGLPVSYDADALGQLVEYMQGDKKTRSGILRFVVLDGLAKPGRLEGP
* :****::**.*** :*:* * ****.*:*:****:****.****: **
Mflv_3756|M.gilvum_PYR-GCK DPSLLAAAYAEVARD------------------
Mvan_2647|M.vanbaalenii_PYR-1 DPSLLAAAYAEVARS------------------
MSMEG_3033|M.smegmatis_MC2_155 DPSLLAAAYAECAGLSPALPQQLSFVKLSPDQL
TH_0108|M.thermoresistible__bu DPSLLTAAYSEIAAQGSKA--------------
MUL_1761|M.ulcerans_Agy99 DPGLLVTAYAGICAP------------------
MMAR_2177|M.marinum_M DPGLLVTAYAGICAP------------------
Mb2567c|M.bovis_AF2122/97 DPGLLVTAYAGVCAP------------------
Rv2538c|M.tuberculosis_H37Rv DPGLLVTAYAGVCAP------------------
MLBr_00518|M.leprae_Br4923 DPGLLVTAYAGLSA-------------------
MAV_3415|M.avium_104 DPSLLAAAYAALADEDAS---------------
MAB_2841c|M.abscessus_ATCC_199 DPSLLAAAYSVVAGEARGEGVLL----------
**.**.:**: .