For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MEPSQGCAEPGRRDVHSQRPGRYGAVASAGLLAAAALGYIGLVDPHGANSAYPACPFKWLTGWDCPACGG LRMVHDLLHGDLVASINDNIFLLVGIPVLGAWALLRRRRGRTSLPLAAMIVVGVAMIVWTVLRNLPGFPL VPAVYGP
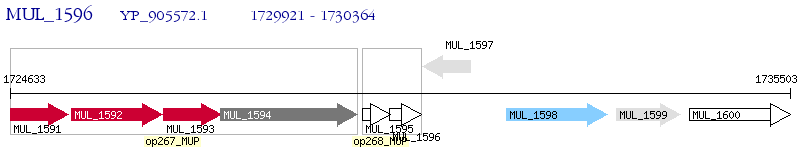
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1596 | - | - | 100% (147) | hypothetical protein MUL_1596 |
| M. ulcerans Agy99 | MUL_3586 | - | 2e-13 | 38.52% (122) | hypothetical protein MUL_3586 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1642 | - | 2e-50 | 70.00% (130) | hypothetical protein Mb1642 |
| M. gilvum PYR-GCK | Mflv_3594 | - | 2e-36 | 52.85% (123) | hypothetical protein Mflv_3594 |
| M. tuberculosis H37Rv | Rv1616 | - | 2e-50 | 70.00% (130) | hypothetical protein Rv1616 |
| M. leprae Br4923 | MLBr_01276 | - | 2e-47 | 59.86% (142) | hypothetical protein MLBr_01276 |
| M. abscessus ATCC 19977 | MAB_3320c | - | 1e-14 | 34.85% (132) | hypothetical protein MAB_3320c |
| M. marinum M | MMAR_2418 | - | 2e-82 | 98.64% (147) | hypothetical protein MMAR_2418 |
| M. avium 104 | MAV_3171 | - | 7e-43 | 66.37% (113) | hypothetical protein MAV_3171 |
| M. smegmatis MC2 155 | MSMEG_3224 | - | 9e-40 | 58.33% (120) | hypothetical protein MSMEG_3224 |
| M. thermoresistible (build 8) | TH_1096 | - | 6e-38 | 55.73% (131) | CONSERVED MEMBRANE PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_2821 | - | 1e-37 | 55.93% (118) | hypothetical protein Mvan_2821 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3594|M.gilvum_PYR-GCK -MAPSPALSTRTRWA---------LGSAAGTVGIGA-LAYIGLGDPHRPG
Mvan_2821|M.vanbaalenii_PYR-1 -MAPSTTTGTRTRLL---------VGASGGAATVGA-LAYIGIGNPHSPS
MSMEG_3224|M.smegmatis_MC2_155 -MEPSERIDHLRTRR---------AALGAGALFAGA-LAYLGFADPHRPG
MUL_1596|M.ulcerans_Agy99 -MEPSQGCAEPGRRDVHSQRPGRYGAVASAGLLAAAALGYIGLVDPHGAN
MMAR_2418|M.marinum_M MMEPSQGSAEPGRRDVHSQRPGRYGAVASAGLLAAA-LGYIGLVDPHGAN
Mb1642|M.bovis_AF2122/97 -MEA-----SGRQRR--------YAAAGSVVLLAGA-LGYIGLVDPHNSN
Rv1616|M.tuberculosis_H37Rv -MEA-----SGRQRR--------YAAAGSVVLLAGA-LGYIGLVDPHNSN
MLBr_01276|M.leprae_Br4923 -MELNQVSRAAQQRRQQDPHRRLYGVAGSGLLLAGA-FGYIGFVDPHNAD
MAV_3171|M.avium_104 ------------------------------MALSGA-LGYIGLADPHDPA
TH_1096|M.thermoresistible__bu -MTSDGATRSRPRRP--GRRHGAVAVTATAVVAALAALTYVALMDPHRPG
MAB_3320c|M.abscessus_ATCC_199 -MTTSRPSTVVSRLA----------APAVVAGAAAAGCAAVWFGDPTTPG
* : : :* .
Mflv_3594|M.gilvum_PYR-GCK F-FPPCPFLEMTGWLCPGCGGLRMTHDLLHGDIAAAFVDNAFILIGVPLL
Mvan_2821|M.vanbaalenii_PYR-1 FVFPSCPFNALTGWLCPACGGLRMTHDLLHGDLAAAFVDNPFVLFGLPLL
MSMEG_3224|M.smegmatis_MC2_155 FLFPPCPFKMLTGWDCPGCGGLRMTHDLLHGDVAAAVVDNVFALVALPVL
MUL_1596|M.ulcerans_Agy99 SAYPACPFKWLTGWDCPACGGLRMVHDLLHGDLVASINDNIFLLVGIPVL
MMAR_2418|M.marinum_M SAYPACPFKWLTGWDCPACGGLRMVHDLLHGDLVASINDNIFLLVGIPVL
Mb1642|M.bovis_AF2122/97 SLYPPCLFKLLTGWNCPACGGLRMIHDLLHGELAASINDNVFLLVGVPVL
Rv1616|M.tuberculosis_H37Rv SLYPPCLFKLLTGWNCPACGGLRMIHDLLHGELAASINDNVFLLVGVPVL
MLBr_01276|M.leprae_Br4923 LVYPLCLFKLLTGWNCPFCGGLRLMHDLLHGDLAASVSDNVFLLVGIPVL
MAV_3171|M.avium_104 SIYPPCPFKWLTGWNCPFCGGLRMTHDLLHGDLWAAVHDNVFLLAAVPTL
TH_1096|M.thermoresistible__bu FLFPACPFHQVTGWFCPGCGGLRMTHHLLHGELTAAAVDNVFLLVGLPVL
MAB_3320c|M.abscessus_ATCC_199 GVLPVCPLKALTGLDCPGCGGLRMAYSLMHGDVLGALRYNAVGLVAAGFL
* * : :** ** *****: : *:**:: .: * . * . *
Mflv_3594|M.gilvum_PYR-GCK IVWALIRRRQ--------SRRVFTVSALAVIFAAVIAWTIVRNLPGFPLV
Mvan_2821|M.vanbaalenii_PYR-1 MGWLLIRRRQ--------GRPAFTAPAYVVMITAVLAWTVVRNLPGFPLV
MSMEG_3224|M.smegmatis_MC2_155 LAWAIVRRRS--------GKPVFTVAVVAVILVSAAVWTVTRNLPGFPLV
MUL_1596|M.ulcerans_Agy99 GAWALLRRRR--------GRTSLPLAAMIVVGVAMIVWTVLRNLPGFPLV
MMAR_2418|M.marinum_M GAWALLRRRR--------GRTSLPLAAMIVVGVAMIVWTVLRNLPGFPLV
Mb1642|M.bovis_AF2122/97 ASWVLLRRRH--------GDLALPIPVMIAVAVAVIAWTVLRNLPGFPLV
Rv1616|M.tuberculosis_H37Rv ASWVLLRRRH--------GDLALPIPVMIAVAVAVIAWTVLRNLPGFPLV
MLBr_01276|M.leprae_Br4923 VGWVVLRRRL--------GQSVLPTVALLTITVASIVWTVLRNVPGFPLV
MAV_3171|M.avium_104 AAFLLVRRAR--------GRRALPAAAVPAVVVATLVWTVLRNLPAFPLF
TH_1096|M.thermoresistible__bu ALWLLIRARL--------RLRLMPPAAVAVLLAAALIWTVVRNLPGFPLI
MAB_3320c|M.abscessus_ATCC_199 AACFVAWTASRWRGESGWNPPWLRGRTPAIIAVVFVAWFVIRLIPVAPFT
: : . : . * : * :* *:
Mflv_3594|M.gilvum_PYR-GCK PTLLTG
Mvan_2821|M.vanbaalenii_PYR-1 PTILDG
MSMEG_3224|M.smegmatis_MC2_155 PTMLPS
MUL_1596|M.ulcerans_Agy99 PAVYGP
MMAR_2418|M.marinum_M PAVYGP
Mb1642|M.bovis_AF2122/97 PTISG-
Rv1616|M.tuberculosis_H37Rv PTISG-
MLBr_01276|M.leprae_Br4923 PTVYSG
MAV_3171|M.avium_104 PTVLGG
TH_1096|M.thermoresistible__bu PTVLSG
MAB_3320c|M.abscessus_ATCC_199 ALRV--
.