For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGDNHPIGPIGAVGAAPGDGVQGGADAIPVRRHQLRAVQRHQVDDFDIQIPAHGSGELRTVTRRSAHSVG ARAGRDRSSGGDNGEAGHGGIISHPCHRHPDVGTGTKLRDDGGVSDEPTGSADPLTTAGETGVTEEPKVS APEPPPRRLRLLLSVAAVVLALDVVTKVLAVKLLPPGQPVSIIGDTVTWTLVRNSGAAFSMATGYTWVLT LIATGVVVGIFWMGRRLVSPWWAVGLGMILGGAMGNLVDRFFRAPGPLRGHVVDFFSVGWWPVFNVADPS VVGGAILLVILSIFGFDFDTVGRRKADER
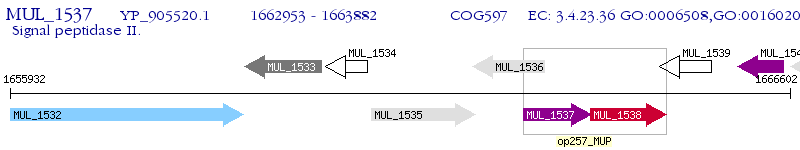
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1537 | lspA | - | 100% (309) | lipoprotein signal peptidase LspA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1566 | lspA | 6e-96 | 88.14% (194) | lipoprotein signal peptidase |
| M. gilvum PYR-GCK | Mflv_3640 | lspA | 2e-87 | 78.57% (196) | lipoprotein signal peptidase |
| M. tuberculosis H37Rv | Rv1539 | lspA | 6e-96 | 88.14% (194) | lipoprotein signal peptidase |
| M. leprae Br4923 | MLBr_01199 | lspA | 2e-84 | 79.08% (196) | lipoprotein signal peptidase |
| M. abscessus ATCC 19977 | MAB_2700c | - | 2e-74 | 83.02% (159) | lipoprotein signal peptidase |
| M. marinum M | MMAR_2361 | lspA | 1e-111 | 100.00% (196) | lipoprotein signal peptidase LspA |
| M. avium 104 | MAV_3231 | lspA | 3e-89 | 92.57% (175) | lipoprotein signal peptidase |
| M. smegmatis MC2 155 | MSMEG_3174 | lspA | 6e-87 | 80.00% (195) | lipoprotein signal peptidase |
| M. thermoresistible (build 8) | TH_1296 | lspA | 2e-77 | 89.19% (148) | PROBABLE LIPOPROTEIN SIGNAL PEPTIDASE LSPA |
| M. vanbaalenii PYR-1 | Mvan_2773 | lspA | 1e-86 | 78.35% (194) | lipoprotein signal peptidase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2700c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3174|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3640|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2773|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1296|M.thermoresistible__bu --------------------------------------------------
MUL_1537|M.ulcerans_Agy99 MGDNHPIGPIGAVGAAPGDGVQGGADAIPVRRHQLRAVQRHQVDDFDIQI
MMAR_2361|M.marinum_M --------------------------------------------------
MAV_3231|M.avium_104 --------------------------------------------------
Mb1566|M.bovis_AF2122/97 --------------------------------------------------
Rv1539|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01199|M.leprae_Br4923 --------------------------------------------------
MAB_2700c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3174|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3640|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2773|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1296|M.thermoresistible__bu --------------------------------------------------
MUL_1537|M.ulcerans_Agy99 PAHGSGELRTVTRRSAHSVGARAGRDRSSGGDNGEAGHGGIISHPCHRHP
MMAR_2361|M.marinum_M --------------------------------------------------
MAV_3231|M.avium_104 --------------------------------------------------
Mb1566|M.bovis_AF2122/97 --------------------------------------------------
Rv1539|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01199|M.leprae_Br4923 --------------------------------------------------
MAB_2700c|M.abscessus_ATCC_199 -------------MSDES-------------------ETARKP--KRRIK
MSMEG_3174|M.smegmatis_MC2_155 -------------MTDETSGPAEPVT-DAPGD----AESPAQP--KRRLR
Mflv_3640|M.gilvum_PYR-GCK ---------MMVGVTDESSSPVEPTTGEGTAE----PGEAVQPPVRRRLR
Mvan_2773|M.vanbaalenii_PYR-1 -------------MTDESSSPVEPVTGEGKAA----GDEPAQPSPPRRLR
TH_1296|M.thermoresistible__bu --------------------------------------------------
MUL_1537|M.ulcerans_Agy99 DVGTGTKLRDDGGVSDEPTGSADPLTTAGETGVTEEPKVSAPEPPPRRLR
MMAR_2361|M.marinum_M -------------MSDEPTGSADPLTTAGETGVTEEPKVSAPEPPPRRLR
MAV_3231|M.avium_104 -------------MPEEPTGSTAPQ-----------------DRPPRRLR
Mb1566|M.bovis_AF2122/97 -------------MPDEPTGSADPLTSTEEAGGAGEPN---APAPPRRLR
Rv1539|M.tuberculosis_H37Rv -------------MPDEPTGSADPLTSTEEAGGAGEPN---APAPPRRLR
MLBr_01199|M.leprae_Br4923 ---------MMGRVPDGPTGLAALVPSVEEAQ---------AMLPPRRLR
MAB_2700c|M.abscessus_ATCC_199 LLLIVASVVLVLDVVTKVLAVRLLQPGKPVPIIGDTVTWTLVRNSGAAFS
MSMEG_3174|M.smegmatis_MC2_155 LLLTVAAVVLFLDVVTKVLAVRLLTPGQPVSIIGDTVTWTLVRNSGAAFS
Mflv_3640|M.gilvum_PYR-GCK LLLGVAAVVLVADIVTKVLAVKLLTPGQPVSIIGDTVTWTLVRNSGAAFS
Mvan_2773|M.vanbaalenii_PYR-1 LLLTVAGVVLVLDIVTKVLAVRLLTPGQPVSIIGDTVTWTLVRNSGAAFS
TH_1296|M.thermoresistible__bu -------VVLVLDIVTKVLAVRFLTPGQPVSIIGDTVTWTLVRNSGAAFS
MUL_1537|M.ulcerans_Agy99 LLLSVAAVVLALDVVTKVLAVKLLPPGQPVSIIGDTVTWTLVRNSGAAFS
MMAR_2361|M.marinum_M LLLSVAAVVLALDVVTKVLAVKLLPPGQPVSIIGDTVTWTLVRNSGAAFS
MAV_3231|M.avium_104 LLLSVAATVLALDIVTKVLAVKLLPPGQPVPIIGDTVTWTLVRNSGAAFS
Mb1566|M.bovis_AF2122/97 MLLSVAVVVLTLDIVTKVVAVQLLPPGQPVSIIGDTVTWTLVRNSGAAFS
Rv1539|M.tuberculosis_H37Rv MLLSVAVVVLTLDIVTKVVAVQLLPPGQPVSIIGDTVTWTLVRNSGAAFS
MLBr_01199|M.leprae_Br4923 LLLSIAAVVLTLDIVTKVLAVKFLLPGKSVSIIGDTVTWTLVRNSGAAFS
.** *:****:**::* **:.*.*******************
MAB_2700c|M.abscessus_ATCC_199 FATGYTWVLTLIAISVVVGILFIGRRLVSTWWAIGLGMILGGALGNLVDR
MSMEG_3174|M.smegmatis_MC2_155 MATGYTWVLTLVATGVVIGIIWMGRRLVSPWWALGLGLILGGATGNLVDR
Mflv_3640|M.gilvum_PYR-GCK MATGYTWVLTLVATGVVIGIVWMGRRLVSPWWALGLGMILGGALGNLVDR
Mvan_2773|M.vanbaalenii_PYR-1 MATGYTWVLTLVATGVVLGIIWMGRRLVSPWWALGLGLILGGAMGNLVDR
TH_1296|M.thermoresistible__bu MATGYTWVLTLIATGVVIGIIWMGRRLVSPWWAIGLGMILGGAMGNLVDR
MUL_1537|M.ulcerans_Agy99 MATGYTWVLTLIATGVVVGIFWMGRRLVSPWWAVGLGMILGGAMGNLVDR
MMAR_2361|M.marinum_M MATGYTWVLTLIATGVVVGIFWMGRRLVSPWWAVGLGMILGGAMGNLVDR
MAV_3231|M.avium_104 MATGYTWVLTLIATGVVVGIFWMGRRLVSPWWAVGLGMILGGAMGNLVDR
Mb1566|M.bovis_AF2122/97 MATGYTWVLTLIATGVVVGIFWMGRRLVSPWWALGLGMILGGAMGNLVDR
Rv1539|M.tuberculosis_H37Rv MATGYTWVLTLIATGVVVGIFWMGRRLVSPWWALGLGMILGGAMGNLVDR
MLBr_01199|M.leprae_Br4923 MATGYTWVLTLIATGVVIGIFWMGRRLVSSWWALGLGMILGGAMGNLVDR
:**********:* .**:**.::******.***:***:***** ******
MAB_2700c|M.abscessus_ATCC_199 FFRGPGPLRGHVVDFLSVGWWPVFNVADPSVVGGAILLVALSLFGYDYDR
MSMEG_3174|M.smegmatis_MC2_155 FFRSPGPLRGHVVDFFSVGWWPVFNVADPSVVGGAILLVALSLFGFDFDT
Mflv_3640|M.gilvum_PYR-GCK FFRSPGPLRGHVVDFLSIGWWPVFNVADPAVVGGAILLVVLSLFGFDFDT
Mvan_2773|M.vanbaalenii_PYR-1 FFRSPGPLRGHVVDFLSIGWWPVFNVADPSVVGGAILLVVLSLFGFDFDS
TH_1296|M.thermoresistible__bu FFRSPGPLRGHVVDFLSIGWWPVFNVADPSVVGGAILLVVLSLLGFDYDT
MUL_1537|M.ulcerans_Agy99 FFRAPGPLRGHVVDFFSVGWWPVFNVADPSVVGGAILLVILSIFGFDFDT
MMAR_2361|M.marinum_M FFRAPGPLRGHVVDFFSVGWWPVFNVADPSVVGGAILLVILSIFGFDFDT
MAV_3231|M.avium_104 FFRAPGPLRGHVVDFLSVGWWPVFNVADPSVVGGAILLVVLSIFGYDFDT
Mb1566|M.bovis_AF2122/97 FFRAPGPLRGHVVDFLSVGWWPVFNVADPSVVGGAILLVILSIFGFDFDT
Rv1539|M.tuberculosis_H37Rv FFRAPGPLRGHVVDFLSVGWWPVFNVADPSVVGGAILLVILSIFGFDFDT
MLBr_01199|M.leprae_Br4923 FFRAPAPLRGHVVDFLSIGWWPVFNVADPSVVVGAILLVVLSIFGFDFDT
***.*.*********:*:***********:** ****** **::*:*:*
MAB_2700c|M.abscessus_ATCC_199 EGR-------------STSKPAS------------SGPEDSVN-ADPQAS
MSMEG_3174|M.smegmatis_MC2_155 VGRRRPGEDAEPSAGASDSTPEAPAADGPDKPAGPVGPEDAAEESKTVGH
Mflv_3640|M.gilvum_PYR-GCK VGRRQPDDADDTVGRRQPDDADDTVGRRQ-----PDDADDTVGRRQPDDA
Mvan_2773|M.vanbaalenii_PYR-1 VGRRQPDGGEDSVGRRQPDGGEDSVGRRQ-----PDGGEDSVGRRRPDGG
TH_1296|M.thermoresistible__bu VGRRRNSGAAG--------DAE----------------------------
MUL_1537|M.ulcerans_Agy99 VGRRK--------------------------------ADER---------
MMAR_2361|M.marinum_M VGRRK--------------------------------ADER---------
MAV_3231|M.avium_104 VGRRKK-------------------------------ADQSRD-------
Mb1566|M.bovis_AF2122/97 VGRRHA-------------------------------DGDTVGRRKADG-
Rv1539|M.tuberculosis_H37Rv VGRRHA-------------------------------DGDTVGRRKADG-
MLBr_01199|M.leprae_Br4923 VGRRKA-------------------------------EFDIAGQRKAEQR
**
MAB_2700c|M.abscessus_ATCC_199 QTKPS-----------------------------------
MSMEG_3174|M.smegmatis_MC2_155 QAEPS-----------------------------------
Mflv_3640|M.gilvum_PYR-GCK DDTVGRRQPDDADDTVGRRQPDDADDTVGRRQPDAESGQG
Mvan_2773|M.vanbaalenii_PYR-1 ED--------------------------------SENG--
TH_1296|M.thermoresistible__bu ----------------------------------------
MUL_1537|M.ulcerans_Agy99 ----------------------------------------
MMAR_2361|M.marinum_M ----------------------------------------
MAV_3231|M.avium_104 ----------------------------------------
Mb1566|M.bovis_AF2122/97 ----------------------------------------
Rv1539|M.tuberculosis_H37Rv ----------------------------------------
MLBr_01199|M.leprae_Br4923 ----------------------------------------