For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAVPAAWAQTHIIDRDGCTALAGRAADDPVLQSAMAAELSTRAMALIAKRGGQHDAIDGSAVQEAAMSFT TSAAFPPLFAQANRVAHDWLFSEAQSAADGERWVVDLAPMLNDSSIQQVLSSYHVTVPDTLRVPLTVSTP QPLHQGQLRRVAVWGPWVSIGAAALAVVCGLLTLITARRRGKALSSLGVSALMVGAAGWAGIEVGGRYVN EALNRTTGDIRRIADVMVNTAEGSLHLWLNLTLLAGVALVVVGAVVAMIRSLWN
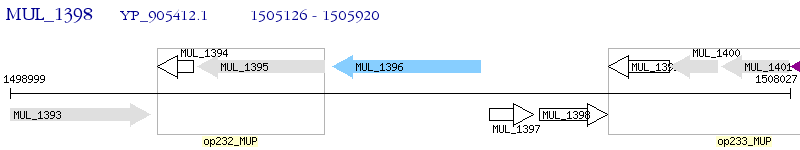
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1398 | - | - | 100% (264) | hypothetical protein MUL_1398 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0131 | - | 3e-71 | 52.09% (263) | hypothetical protein Mflv_0131 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4133c | - | 8e-48 | 38.95% (267) | hypothetical protein MAB_4133c |
| M. marinum M | MMAR_0764 | - | 1e-146 | 99.24% (264) | hypothetical protein MMAR_0764 |
| M. avium 104 | MAV_4699 | - | 3e-86 | 61.83% (262) | hypothetical protein MAV_4699 |
| M. smegmatis MC2 155 | MSMEG_0892 | - | 5e-74 | 52.87% (261) | hypothetical protein MSMEG_0892 |
| M. thermoresistible (build 8) | TH_2840 | - | 1e-64 | 49.22% (258) | PUTATIVE putative conserved hypothetical protein |
| M. vanbaalenii PYR-1 | Mvan_0773 | - | 2e-72 | 53.05% (262) | hypothetical protein Mvan_0773 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0131|M.gilvum_PYR-GCK MRFLAALLLWLLTTMALAVAIPAVWTQWNVIGEDGYAGLAASAARDSRLQ
Mvan_0773|M.vanbaalenii_PYR-1 MRFFAALLLWLLTTIALAAAVPTVWAQTHIVGEDGYAALAASAARDGRLQ
TH_2840|M.thermoresistible__bu VRFLTTLLLWLVTTVALAATIPALWAQHHIVDEHGYSDLAAAAARDPVLQ
MUL_1398|M.ulcerans_Agy99 ------------------MAVPAAWAQTHIIDRDGCTALAGRAADDPVLQ
MMAR_0764|M.marinum_M MRLLATVVLWLATTVALAVAVPAAWAQTHIIDRDGYTALAGRAADDPVLQ
MAV_4699|M.avium_104 MRFAGVVLGWLIATLALAVAVPAGWVQLHVVDADGYAALARRAAADPALQ
MSMEG_0892|M.smegmatis_MC2_155 MRFLMTLFLWLLTTVLLAAAVPSAWAQKNVIDVAGYSAFAEAAAKDPQLQ
MAB_4133c|M.abscessus_ATCC_199 MRFLFAALLWLLTTAALAVTIAAAWAQSRLVDENGYAALTAPAAADPRVQ
::.: *.* .::. * : :: ** * :*
Mflv_0131|M.gilvum_PYR-GCK DAMAGELTTQVLALATDNGYS---LNPGLAREVADAYTRNDGFPGQFAQA
Mvan_0773|M.vanbaalenii_PYR-1 EAMAAELTTQVVSLGADNGYS---LNPDLVRRVADSYTRNSGFPGQFAQA
TH_2840|M.thermoresistible__bu EAMADELADQFTAWAADRGYR---LNRSMVGDVARAYTGNPGFPAQFAQA
MUL_1398|M.ulcerans_Agy99 SAMAAELSTRAMALIAKRGGQHDAIDGSAVQEAAMSFTTSAAFPPLFAQA
MMAR_0764|M.marinum_M SAMAAELSTRAMALIAKRGGQHDAIDGSAVQEAAMSFTTSAAFPPLFAQA
MAV_4699|M.avium_104 SAMAAELTTRAMALIAEHGGGRYPVDGTQVHDAAAAFTAGPAFPPLFAQA
MSMEG_0892|M.smegmatis_MC2_155 QAVAGELATQVSTLVRDNGGE---VNADMVRGTANAYTAGEDFPGQFARA
MAB_4133c|M.abscessus_ATCC_199 HAIAGELTTQIVSLGKKQGSR---VDESQVAELVASYTSGPQFEADFGSV
*:* **: : : ..* :: . . ::* . * *. .
Mflv_0131|M.gilvum_PYR-GCK NRIAHRWMFTGAVPSGNST-DEWLIDVAPMLSDPSFAATLGTLDLQVPPT
Mvan_0773|M.vanbaalenii_PYR-1 NRIAHRWMFTGAVPSGNST-DEWLIDVAPMLSDPSFSATLGTLDLQIPPT
TH_2840|M.thermoresistible__bu NRIAHRWMFTRSTPQGEST-DQWLIDIAPMLSDPSFRDTAGDLDLEVPDT
MUL_1398|M.ulcerans_Agy99 NRVAHDWLFSEAQ-SAADG-ERWVVDLAPMLNDSSIQQVLSSYHVTVPDT
MMAR_0764|M.marinum_M NRVAHDWLFSEAQ-SAADG-ERWVVDLAPMLNDSSIQQVLSSYHVTVPDT
MAV_4699|M.avium_104 NRAAHAWLFDG---SGAGG-DQWAVDVAPMLDDPSIRPLLRRHNVAVPAK
MSMEG_0892|M.smegmatis_MC2_155 NEIAHTWLFTDRI-RRDEG-GSWVIDLAPMLADSSFEDTLSNFNVAVPQT
MAB_4133c|M.abscessus_ATCC_199 NRMAHQWLFTNSSSAQRDSQGGWQIDIAPMIKSLAPQG----LSIKAPET
*. ** *:* * :*:***: . : : * .
Mflv_0131|M.gilvum_PYR-GCK LTVPITVDTPE-LQPGKLRWLATWGPWAAVGAAAITGVFALLTLAAARSR
Mvan_0773|M.vanbaalenii_PYR-1 LTVPITVDSPE-LQPGKLRALATWGPWVSIGAAVVTGMFGLLTLAAARSR
TH_2840|M.thermoresistible__bu LLVPITVPTDS-LRPGRLATVATWGPWVTIAAATLTTMLAMLTVAAARAR
MUL_1398|M.ulcerans_Agy99 LRVPLTVSTPQPLHQGQLRRVAVWGPWVSIGAAALAVVCGLLTLITARRR
MMAR_0764|M.marinum_M LRVPLTVSTPQPLHQGQLRRVAVWGPWVSIGAAALAVVCGLLTLITARRR
MAV_4699|M.avium_104 LTVPLTVSVPR---QGRLSGLTRWGPWLSLGSAALCGVGALLTLAAARRR
MSMEG_0892|M.smegmatis_MC2_155 LTVPIASEPPSGLEPGRLRPLATWGPWVSVGTTVLAGVFALLTLATARSR
MAB_4133c|M.abscessus_ATCC_199 LKVSVTDDKLAGLTPGRLVPVARFGQLAVWIFFGLTLVLAGLTQLAVRSR
* *.:: *:* :: :* : : . ** :.* *
Mflv_0131|M.gilvum_PYR-GCK GKALTALGVSALLVGAAGWTGIEMGRRFVEAGLNRTTGDIRTVAEVMVAH
Mvan_0773|M.vanbaalenii_PYR-1 GKALTALGVSALLVGAAGWAAIEVGGRFVDAGLDRTTGNIRTVAEVMVAS
TH_2840|M.thermoresistible__bu GKAVAALGVSALLVGGTGWAAVEVLRGRIDEALNRTETGVRQVAEVMVGQ
MUL_1398|M.ulcerans_Agy99 GKALSSLGVSALMVGAAGWAGIEVGGRYVNEALNRTTGDIRRIADVMVNT
MMAR_0764|M.marinum_M GKALSSLGVSALMVGAAGWAGIEVGGRYVNEALNRTTGDIRRIADVMVNT
MAV_4699|M.avium_104 GKALSSLGVAALLVGAAGWAGLEAAGRYVNDALNHTTGNVRRIAEVMVAH
MSMEG_0892|M.smegmatis_MC2_155 GKTLAALGISALLVGAAGWAGLEIARRYLNDALNHTSGDIRAIADAMVGH
MAB_4133c|M.abscessus_ATCC_199 AKGLAGLGISALLVGAAGWAGLEIGRSYLDRPLSRVTGNVRDVADSLVTA
.* ::.**::**:**.:**:.:* :: *.:. .:* :*: :*
Mflv_0131|M.gilvum_PYR-GCK AESSMHLWLNYTLLAGVGLVVIGALGAVLGSVLRRPSPSVGS
Mvan_0773|M.vanbaalenii_PYR-1 AEASLHQWLNYTLIAGGALVLLGVTAAVLGSVLRGPDRA---
TH_2840|M.thermoresistible__bu AVDSLHHWLNLTLAAGGALVLLGVVVAMLGAVLRG-DRV---
MUL_1398|M.ulcerans_Agy99 AEGSLHLWLNLTLLAGVALVVVGAVVAMIRSLWN--------
MMAR_0764|M.marinum_M AEGSLHLWLNLTLLAGVALVVVGAVAAMIRSLWN--------
MAV_4699|M.avium_104 AESGVHQWLNATLLAGAALAGLGVLVAVLGGLAGAASRR---
MSMEG_0892|M.smegmatis_MC2_155 GIGSLHQWLNLTLAVGGGLVVLGVVVAMLGGLRRSR------
MAB_4133c|M.abscessus_ATCC_199 AVGNAHHWLSLTMAAGGLVIVIGVVSGLLGALLRR-------
. . * **. *: .* : :*. .:: .: