For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTPAARRRRSRVVGRDAGSEAIRPICRLAGRGYGATVPGVVEYVGTCPFVGQVIRANKANKHSPEGSRG SDMDGAMARAIRAGDDSEIADGLTRREHDILAFERQWWKFAGVKEDAIKELFSMSATRYYQVLNALVDRP EALAADPMLVKRLRRLRASRQKARAARRLGFEVT
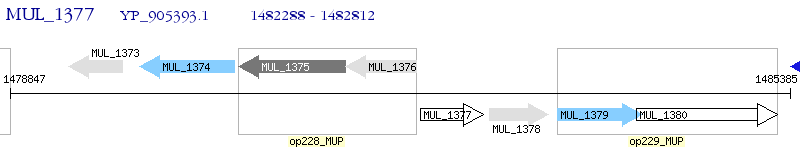
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1377 | - | - | 100% (174) | hypothetical protein MUL_1377 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0438 | - | 2e-51 | 95.10% (102) | hypothetical protein Mb0438 |
| M. gilvum PYR-GCK | Mflv_0174 | - | 6e-49 | 92.16% (102) | Fis family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0430 | - | 2e-51 | 95.10% (102) | hypothetical protein Rv0430 |
| M. leprae Br4923 | MLBr_01927 | - | 8e-51 | 95.10% (102) | hypothetical protein MLBr_01927 |
| M. abscessus ATCC 19977 | MAB_4186c | - | 5e-43 | 79.13% (115) | hypothetical protein MAB_4186c |
| M. marinum M | MMAR_0745 | - | 2e-52 | 99.02% (102) | hypothetical protein MMAR_0745 |
| M. avium 104 | MAV_4724 | - | 4e-51 | 96.08% (102) | hypothetical protein MAV_4724 |
| M. smegmatis MC2 155 | MSMEG_0833 | - | 4e-48 | 88.24% (102) | hypothetical protein MSMEG_0833 |
| M. thermoresistible (build 8) | TH_0950 | - | 4e-48 | 89.22% (102) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_0733 | - | 1e-48 | 91.18% (102) | Fis family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_1377|M.ulcerans_Agy99 MTTPAARRRRSRVVGRDAGSEAIRPICRLAGRGYGATVPGVVEYVGTCPF
MMAR_0745|M.marinum_M --------------------------------------------------
MLBr_01927|M.leprae_Br4923 --------------------------------------------------
Mb0438|M.bovis_AF2122/97 --------------------------------------------------
Rv0430|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4724|M.avium_104 --------------------------------------------------
Mflv_0174|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0733|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0833|M.smegmatis_MC2_155 --------------------------------------------------
TH_0950|M.thermoresistible__bu --------------------------------------------------
MAB_4186c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_1377|M.ulcerans_Agy99 VGQVIRANKANKHSPEGSRGSDMDGAMARAIRA-----GDDS--------
MMAR_0745|M.marinum_M ----------------------MDGAMARANRA-----GDDS--------
MLBr_01927|M.leprae_Br4923 ----------------------MDGAMARAHRA-----GDDV--------
Mb0438|M.bovis_AF2122/97 ----------------------MDSAMARAIRS-----GDDA--------
Rv0430|M.tuberculosis_H37Rv ----------------------MDSAMARAIRS-----GDDA--------
MAV_4724|M.avium_104 ----------------------MDSAMARANRS-----GDDA--------
Mflv_0174|M.gilvum_PYR-GCK ----------------------MDGAIARAERS-----GDDS--------
Mvan_0733|M.vanbaalenii_PYR-1 ----------------------MDGAIARAERS-----GDDT--------
MSMEG_0833|M.smegmatis_MC2_155 ----------------------MDGAMARTEQS-----GDDS--------
TH_0950|M.thermoresistible__bu ----------------------MDGAIARTEQS-----GDDS--------
MAB_4186c|M.abscessus_ATCC_199 ----------------------MDGAQARAEQSKAANAGEDSPAVGSDGV
**.* **: :: *:*
MUL_1377|M.ulcerans_Agy99 EIADGLTRREHDILAFERQWWKFAGVKEDAIKELFSMSATRYYQVLNALV
MMAR_0745|M.marinum_M EIADGLTRREHDILAFERQWWKFAGVKEDAIKELFSMSATRYYQVLNALV
MLBr_01927|M.leprae_Br4923 EIVDGLTRREHDILAFERQWWKFAGVKEEAIKELFSMSATRYYQVLNALV
Mb0438|M.bovis_AF2122/97 EVADGLTRREHDILAFERQWWKFAGVKEEAIKELFSMSATRYYQVLNALV
Rv0430|M.tuberculosis_H37Rv EVADGLTRREHDILAFERQWWKFAGVKEEAIKELFSMSATRYYQVLNALV
MAV_4724|M.avium_104 EIADGLTRREHDILAFERQWWKFAGVKEDAIKELFSMSATRYYQVLNALV
Mflv_0174|M.gilvum_PYR-GCK EPTDGLTRREHDILAFERQWWKYAGSKEDAIKELFSMSATRYYQVLNALV
Mvan_0733|M.vanbaalenii_PYR-1 EPTDGLTRREHDILAFERQWWKYAGSKEDAIKELFSMSATRYYQVLNALV
MSMEG_0833|M.smegmatis_MC2_155 DLTDGLTRREHDILAFERQWWKYAGSKEDAIKELFSMSATRYYQVLNALV
TH_0950|M.thermoresistible__bu ELTDGLTRREHEILAFERQWWKYAGSKEEAIKELFSMSATRYYQVLNALV
MAB_4186c|M.abscessus_ATCC_199 EIAAGLSRREHDILAFERQWWKYAGSKEDAIKELFGMSATRYYQVLNALV
: . **:****:**********:** **:******.**************
MUL_1377|M.ulcerans_Agy99 DRPEALAADPMLVKRLRRLRASRQKARAARRLGFEVT-
MMAR_0745|M.marinum_M DRPEALAADPMLVKRLRRLRASRQKARAARRLGFEVT-
MLBr_01927|M.leprae_Br4923 DRPEALAVDPMLVKRLRRLRASRQKARAARRLGFEVT-
Mb0438|M.bovis_AF2122/97 DRPEALAADPMLVKRLRRLRASRQKARAARRLGFEVT-
Rv0430|M.tuberculosis_H37Rv DRPEALAADPMLVKRLRRLRASRQKARAARRLGFEVT-
MAV_4724|M.avium_104 DRPEALAADPMLVKRLRRLRASRQKARAARRLGFEVT-
Mflv_0174|M.gilvum_PYR-GCK DRPEALAADPMLVKRLRRLRASRQKARAARRLGFDVT-
Mvan_0733|M.vanbaalenii_PYR-1 DRPEALAADPMLVKRLRRLRASRQKARAARRLGFDVT-
MSMEG_0833|M.smegmatis_MC2_155 DRPEALAADPMLVKRLRRLRASRQKARAARRLGFDISK
TH_0950|M.thermoresistible__bu DRPEALAADPMLVKRLRRLRASRQKARAARRLGFEVT-
MAB_4186c|M.abscessus_ATCC_199 DRQEALAADPMLVKRLRRMRASRQKARAARRLGFEVT-
** ****.**********:***************:::