For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVEIGMGRTARRTYELSEISIVPSRRTRSSKDVSTAWQLDAYRFEIPVVAHPTDALVSPEFAIELGRLGG LAVLNGEGLIGRHADVAAKITQLVEAASKEPEPTAAIRLLQELHAAPLDPGLLGAAVARIREAGVTTAVR VSPQNAQALTPVLVQAGIDLLVIQGTIVSAEHVSSDGEPLNLKTFISELDIPVVAGGVLDHRTALHLMRT GAAGVIVGYGSTKGMTSSDEVLGISVPMATAIADAAAARREYLDETGGRYVHVLADGDIHTSGELAKAIA CGADAVVLGTPLAESAEALGEGWFWPAAAAHPSLPRGALLQIAMGERPSLERVLNGPSDDPFGTLNLVGG LRRSMVKAGYCDLKEFQKVGLAVGG
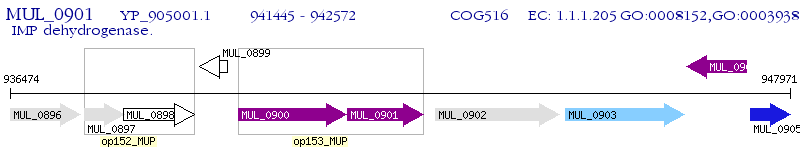
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0901 | guaB3 | - | 100% (375) | inosine 5-monophosphate dehydrogenase |
| M. ulcerans Agy99 | MUL_0900 | guaB2 | 1e-12 | 35.33% (150) | inosine 5'-monophosphate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3444c | guaB3 | 0.0 | 92.53% (375) | inosine 5-monophosphate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4908 | - | 0.0 | 87.30% (378) | inositol-5-monophosphate dehydrogenase |
| M. tuberculosis H37Rv | Rv3410c | guaB3 | 0.0 | 92.53% (375) | inosine 5-monophosphate dehydrogenase |
| M. leprae Br4923 | MLBr_00388 | guaB3 | 0.0 | 89.95% (368) | inosine 5-monophosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3720c | - | 1e-180 | 83.38% (373) | inosine 5-monophosphate dehydrogenase |
| M. marinum M | MMAR_1138 | guaB3 | 0.0 | 99.47% (375) | inosine-5'-monophosphate (imp) dehydrogenase, GuaB3 |
| M. avium 104 | MAV_4355 | - | 0.0 | 90.48% (378) | inositol-5-monophosphate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1603 | - | 0.0 | 90.37% (374) | inositol-5-monophosphate dehydrogenase |
| M. thermoresistible (build 8) | TH_4385 | - | 1e-17 | 92.86% (42) | PUTATIVE - |
| M. vanbaalenii PYR-1 | Mvan_1512 | - | 0.0 | 86.63% (374) | inositol-5-monophosphate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0901|M.ulcerans_Agy99 ---MVEIGMGRTARRTYELSEISIVPSRRTRSSKDVSTAWQLDAYRFEIP
MMAR_1138|M.marinum_M ---MVEIGMGRTARRTYELSEISIVPSRRTRSSKDVSTAWQLDAYRFEIP
Mb3444c|M.bovis_AF2122/97 ---MVEIGMGRTARRTYELSEISIVPSRRTRSSKDVSTAWQLDAYRFEIP
Rv3410c|M.tuberculosis_H37Rv ---MVEIGMGRTARRTYELSEISIVPSRRTRSSKDVSTAWQLDAYRFEIP
MLBr_00388|M.leprae_Br4923 --------MGRVARRTYELREISIVPSRRTRSSKDVSTAWQLDAYRFEIP
Mflv_4908|M.gilvum_PYR-GCK MRDMVEIGMGRTARRTYELGDINIVPSRRTRSSKDVSTAWQLDAYRFEIP
Mvan_1512|M.vanbaalenii_PYR-1 MRDMVEIGMGRTARRTYELDDINIVPSRRTRSSKDVSTAWQLDAYRFEIP
MSMEG_1603|M.smegmatis_MC2_155 ---MVEIGMGRTARRTYELEDVTIVPSRRTRSSKDVSTAWQLDAYRFEIP
MAV_4355|M.avium_104 ---MVEIGMGRTARRSYELSEVSIVASRRTRSSQDVSTAWQLDAYRFEIP
TH_4385|M.thermoresistible__bu --------------------------------------------------
MAB_3720c|M.abscessus_ATCC_199 MRDLVEIGMGRTARRTYELDDVNIVPSRRTRSSQDVSTAWQLDAYRFEIP
MUL_0901|M.ulcerans_Agy99 VVAHPTDALVSPEFAIELGRLGGLAVLNGEGLIGRHADVAAKITQLVEAA
MMAR_1138|M.marinum_M VVAHPTDALVSPEFAIELGRLGGLAVLNGEGLIGRHADVAAKITQLVEAA
Mb3444c|M.bovis_AF2122/97 VVAHPTDALVSPEFAIELGRLGGLGVLNGEGLIGRHLDVEAKIAQLLEAA
Rv3410c|M.tuberculosis_H37Rv VVAHPTDALVSPEFAIELGRLGGLGVLNGEGLIGRHLDVEAKIAQLLEAA
MLBr_00388|M.leprae_Br4923 VVAHPTDALVSPEFAIELGRLGGLGVLNGEGLIGRHADVGAKVAQLIEAA
Mflv_4908|M.gilvum_PYR-GCK VLAHPTDALVSVEFAIELGRLGGLGVLNGEGLIGRHADVEDKIARVVEVA
Mvan_1512|M.vanbaalenii_PYR-1 VVAHPTDALVSVEFAIEMGRCGGLGVLNGEGLIGRHADVADKIAQVIEVA
MSMEG_1603|M.smegmatis_MC2_155 VIAHPTDALVSPEFAIEMGRLGGLGVLNGEGLIGRHADVEEKIAQVVEVA
MAV_4355|M.avium_104 VLAHPTDALVSPEFAIELGRLGGLGVLNGEGLIGRHADVEAKIAQLVEAA
TH_4385|M.thermoresistible__bu --------------------------------------------------
MAB_3720c|M.abscessus_ATCC_199 VVAHPTDALVSPRFAIELGKAGGLGVINGEGLWGRHADVEARIAEVVEVA
MUL_0901|M.ulcerans_Agy99 SKEPEPTAAIRLLQELHAAPLDPGLLGAAVARIREAGVTTAVRVSPQNAQ
MMAR_1138|M.marinum_M SKEPEPTAAIRLLQELHAAPLDPGLLGAAVARIREAGVTTAVRVSPQNAQ
Mb3444c|M.bovis_AF2122/97 AADPEPSTAIRLLQELHAAPLNPDLLGAAVARIREAGVTTAVRVSPQNAQ
Rv3410c|M.tuberculosis_H37Rv AADPEPSTAIRLLQELHAAPLNPDLLGAAVARIREAGVTTAVRVSPQNAQ
MLBr_00388|M.leprae_Br4923 EKEPEPATAVCLLQELHAAPLNPDLLGSAVARIREAGVTTAVRVSPQNAQ
Mflv_4908|M.gilvum_PYR-GCK EKDPDPAAATRLLQQLHAAPLDPELLGAAVSRIREAGVTTAVRVSPQNAQ
Mvan_1512|M.vanbaalenii_PYR-1 VKDPDPSASTRLLQQLHSAPLDLDLLGSAVARIRDAGVTTAVRVSPQNAQ
MSMEG_1603|M.smegmatis_MC2_155 AKEPEPSAAIRLLQQLHAAPLDPDLLGAAVARIREAGVTTAVRVSPQNAQ
MAV_4355|M.avium_104 SKEPEPSAAIRLLQELHAAPLNPDLLGSAVARIREAGVTTAVRVSPQNAQ
TH_4385|M.thermoresistible__bu --------------------------------------------------
MAB_3720c|M.abscessus_ATCC_199 QKEPEPSAAIRLLQQLHSAPIDPDLLAAAIAEVRGAGVTTAVRVSPQNAQ
MUL_0901|M.ulcerans_Agy99 ALTPVLVQAGIDLLVIQGTIVSAEHVSSD----GEPLNLKTFISELDIPV
MMAR_1138|M.marinum_M ALTPVLVQAGIDLLVIQGTIVSAEHVSSD----GEPLNLKTFISELDIPV
Mb3444c|M.bovis_AF2122/97 WLTPVLVAAGIDLLVIQGTIVSAERVASD----GEPLNLKTFISELDIPV
Rv3410c|M.tuberculosis_H37Rv WLTPVLVAAGIDLLVIQGTIVSAERVASD----GEPLNLKTFISELDIPV
MLBr_00388|M.leprae_Br4923 VLTPVLLAAGIDLLVVQGSIVSAECVASG----GEPLNLKTFISELDVPV
Mflv_4908|M.gilvum_PYR-GCK ALTPTLVAAGIDLLVIQGTIISAERVAKDHDGEGEPLNLKTFISELDVPV
Mvan_1512|M.vanbaalenii_PYR-1 ALTPTLVAAGIDLLVIQGTIISAERVAND----GEPLNLKTFISELDVPV
MSMEG_1603|M.smegmatis_MC2_155 ALTPTLVAAGIDLLVIQGTIISAERVASD----GEPLNLKTFISELDVPV
MAV_4355|M.avium_104 ALTPVLLQAGIDLLVIQGTIVSAERVALERDGAGEPLNLKTFISELDVPV
TH_4385|M.thermoresistible__bu --------------------------------------------------
MAB_3720c|M.abscessus_ATCC_199 LLTPHLIAAGIDLLVIHGTIVSAERVARD----GEPLNLKTFISELDIPV
MUL_0901|M.ulcerans_Agy99 VAGGVLDHRTALHLMRTGAAGVIVGYGSTKGMTSSDEVLGISVPMATAIA
MMAR_1138|M.marinum_M VAGGVLDHRTALHLMRTGAAGVIVGYGSTEGMTSSDEVLGISVPMATAIA
Mb3444c|M.bovis_AF2122/97 VAGGVLDHRTALHLMRTGAAGVIVGYGSTQGVTTTDEVLGISVPMATAIA
Rv3410c|M.tuberculosis_H37Rv VAGGVLDHRTALHLMRTGAAGVIVGYGSTQGVTTTDEVLGISVPMATAIA
MLBr_00388|M.leprae_Br4923 VAGGVLDHRTALHLMRTGAAGVIVGYGSTRGATTSDEVLGISVPMATAIA
Mflv_4908|M.gilvum_PYR-GCK VAGGVLDHRTALHLMRTGAAGVIVGYGSTSGVTTSDEVLGISVPMATAIA
Mvan_1512|M.vanbaalenii_PYR-1 VAGGVLDHRTALHLMRTGAAGVIVGYGSTAGVTTSDEVLGISVPMATAIA
MSMEG_1603|M.smegmatis_MC2_155 VAGGVLDHRTALHLMRTGAAGVIVGYGSTSGVTTSDEVLGISVPMATAIA
MAV_4355|M.avium_104 VAGGVQDHRTALHLMRTGAAGVIVGYGATRGVTTSDEVLGISVPMATAIA
TH_4385|M.thermoresistible__bu --------------------------------------------------
MAB_3720c|M.abscessus_ATCC_199 VAGGVIDHRTALHLMRTGAAGVIVGYGQTAGATTSSEVLGVSVAMATAIA
MUL_0901|M.ulcerans_Agy99 DAAAARREYLDETGGRYVHVLADGDIHTSGELAKAIACGADAVVLGTPLA
MMAR_1138|M.marinum_M DAAAARREYLDETGGRYVHVLADGDIHTSGELAKAIACGADAVVLGTPLA
Mb3444c|M.bovis_AF2122/97 DAAAARRDYLDETGGRYVHVLADGDIHTSGELAKAIACGADAVVLGTPLA
Rv3410c|M.tuberculosis_H37Rv DAAAARRDYLDETGGRYVHVLADGDIHTSGELAKAIACGADAVVLGTPLA
MLBr_00388|M.leprae_Br4923 DAAAARREYLDETGGRYVHVLADGDIYTSGELAKAIACGADAVVLGTPLA
Mflv_4908|M.gilvum_PYR-GCK DAAAARREYLDETGGRYVHVLADGDIHTSGDLAKAIACGADAVVLGTPLA
Mvan_1512|M.vanbaalenii_PYR-1 DAAAARREYLDETGGRYVHVLADGDIHTSGDLAKAIACGADAVVLGTPLA
MSMEG_1603|M.smegmatis_MC2_155 DAAAARREYLDETGGRYVHVLADGDIHSSGDLAKAIACGADAVVLGTPLA
MAV_4355|M.avium_104 DAAAARREYLDETGGRYVHVLADGDIHTSGELAKAIACGADAVVLGTPLA
TH_4385|M.thermoresistible__bu --------------------------------------------------
MAB_3720c|M.abscessus_ATCC_199 DAAAARREYLDETGGRYVHVLADGDIHTSGDLAKSIACGADAVVLGTPLA
MUL_0901|M.ulcerans_Agy99 ESAEALGEGWFWPAAAAHPSLPRGALLQIAMGERPSLERVLNGPSDDPFG
MMAR_1138|M.marinum_M ESAEALGEGWFWPAAAAHPSLPRGALLQIAMGERPSLERVLNGPSDDPFG
Mb3444c|M.bovis_AF2122/97 ESAEALGEGWFWPAAAAHPSLPRGALLQIAVGERPPLARVLGGPSDDPFG
Rv3410c|M.tuberculosis_H37Rv ESAEALGEGWFWPAAAAHPSLPRGALLQIAVGERPPLARVLGGPSDDPFG
MLBr_00388|M.leprae_Br4923 QSAEALGGGWFWPAAAAHPSLPRGALLQTAVGERPSLQQVLNGPSDNPFG
Mflv_4908|M.gilvum_PYR-GCK ISAEALGGGWFWPAAAAHPSLPRGSLLQVVDEERPSLAQVLNGPSDDPFG
Mvan_1512|M.vanbaalenii_PYR-1 VSAESLGGGWFWPAAAAHPSLPRGAMMQVADEERPALSQVLNGPSDDPFG
MSMEG_1603|M.smegmatis_MC2_155 TSAEALGNGWFWPAAAAHPSLPRGALLQVALGERPSLEQVLTGPSDDPFG
MAV_4355|M.avium_104 EAAEALGEGWFWPAAAAHPSLPRGALLQVAIGERPPLRQVLNGPSDDPFG
TH_4385|M.thermoresistible__bu ---------------------------------------VLNGPSDDPFG
MAB_3720c|M.abscessus_ATCC_199 AAQEAAGDGWFWPNAAAHPSLPRGALLQVAVGERPPLSEVLAGPSDDPFG
** ****:***
MUL_0901|M.ulcerans_Agy99 TLNLVGGLRRSMVKAGYCDLKEFQKVGLAVGG
MMAR_1138|M.marinum_M TLNLVGGLRRSMAKAGYCDLKEFQKVGLAVGG
Mb3444c|M.bovis_AF2122/97 GLNLVGGLRRSMAKAGYCDLKEFQKVGLTVGG
Rv3410c|M.tuberculosis_H37Rv GLNLVGGLRRSMAKAGYCDLKEFQKVGLTVGG
MLBr_00388|M.leprae_Br4923 TLNLVGGLRRSMAKAGYCDLKEFQKVGLIVSI
Mflv_4908|M.gilvum_PYR-GCK SLNLVGGLARSMAKAGYCDLKEFQKVGLTVGC
Mvan_1512|M.vanbaalenii_PYR-1 SLNLVGGLARSMAKAGYCDLKEFQKVGLTVGR
MSMEG_1603|M.smegmatis_MC2_155 SLNLVGGLRRSMAKAGYCDLKEFQKVGLTVGS
MAV_4355|M.avium_104 SLNLVGGLRRSMAKAGYCDLKEFQKVGLTVGS
TH_4385|M.thermoresistible__bu SLNLVGGLRRSMAKAGYCDLKEFQKVGLTVGS
MAB_3720c|M.abscessus_ATCC_199 TLNLVGGLRRSMAKSGYCDLKEFQKVGLTVNS
******* ***.*:************* *.