For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MARYTGPVTRKSRRLGTDLVGGDQSFEKRPYPPGQHGRARIKDSEYRQQLQEKQKARFTYGVMEKQFRRY YEEAVRHSGKTGEELLKILESRLDNVVYRAGLARTRRMARQLVSHGHFSVNGVHVNVPSYRVSQYDIIDV RDNSLNTVPFQIARETAGDRPIPSWLQVVGGRQRILIHQLPERAQIEVPLTEQLIVEYYSK
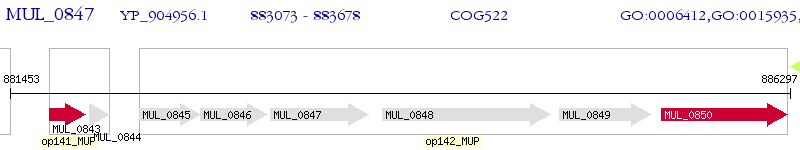
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0847 | rpsD | - | 100% (201) | 30S ribosomal protein S4 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3487c | rpsD | 1e-106 | 92.54% (201) | 30S ribosomal protein S4 |
| M. gilvum PYR-GCK | Mflv_4971 | rpsD | 1e-101 | 87.56% (201) | 30S ribosomal protein S4 |
| M. tuberculosis H37Rv | Rv3458c | rpsD | 1e-106 | 92.54% (201) | 30S ribosomal protein S4 |
| M. leprae Br4923 | MLBr_01958 | rpsD | 1e-104 | 91.04% (201) | 30S ribosomal protein S4 |
| M. abscessus ATCC 19977 | MAB_3771c | rpsD | 1e-100 | 86.07% (201) | 30S ribosomal protein S4 |
| M. marinum M | MMAR_1089 | rpsD | 1e-113 | 99.50% (201) | 30S ribosomal protein S4, RpsD |
| M. avium 104 | MAV_4399 | rpsD | 1e-103 | 90.55% (201) | 30S ribosomal protein S4 |
| M. smegmatis MC2 155 | MSMEG_1523 | rpsD | 1e-101 | 88.56% (201) | 30S ribosomal protein S4 |
| M. thermoresistible (build 8) | TH_0031 | rpsD | 5e-98 | 85.57% (201) | PROBABLE 30S RIBOSOMAL PROTEIN S4 RPSD |
| M. vanbaalenii PYR-1 | Mvan_1435 | rpsD | 1e-101 | 87.56% (201) | 30S ribosomal protein S4 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0847|M.ulcerans_Agy99 MARYTGPVTRKSRRLGTDLVGGDQSFEKRPYPPGQHGRARIKDSEYRQQL
MMAR_1089|M.marinum_M MARYTGPVTRKSRRLGTDLVGGDQSFEKRPYPPGQHGRARIKDSEYRQQL
Mb3487c|M.bovis_AF2122/97 MARYTGPVTRKSRRLRTDLVGGDQAFEKRPYPPGQHGRARIKESEYLLQL
Rv3458c|M.tuberculosis_H37Rv MARYTGPVTRKSRRLRTDLVGGDQAFEKRPYPPGQHGRARIKESEYLLQL
MLBr_01958|M.leprae_Br4923 MARYTGPITRKSRRLRIDLVGGDQAFEKRPYPPGQHGRARIKESEYLLQL
MAV_4399|M.avium_104 MARYTGPVTRKSRRLRTDLVGGDQAFEKRPYPPGQHGRARIKESEYLLQL
TH_0031|M.thermoresistible__bu MARYTGPVTRKSRRLRVDLVGGDQAFEKRPYPPGQHGRARIKESEYLLQL
Mflv_4971|M.gilvum_PYR-GCK MARYTGPVTRKSRRLGVDLVGGDQSFEKRPYPPGQHGRARIKESEYRTQL
Mvan_1435|M.vanbaalenii_PYR-1 MARYTGPVTRKSRRLGVDLVGGDQSFEKRPYPPGQHGRARIKESEYRTQL
MSMEG_1523|M.smegmatis_MC2_155 MARYTGPATRKSRRLGVDLVGGDQSFEKRPYPPGQHGRARIKESEYRQQL
MAB_3771c|M.abscessus_ATCC_199 MARYTGPVTRKSRRLGVDLVGGSSAYEKRPYPPGQHGRARIKESEYRQQL
******* ******* *****..::****************:*** **
MUL_0847|M.ulcerans_Agy99 QEKQKARFTYGVMEKQFRRYYEEAVRHSGKTGEELLKILESRLDNVVYRA
MMAR_1089|M.marinum_M QEKQKARFTYGVMEKQFRRYYEEAVRHSGKTGEELLKILESRLDNVVYRA
Mb3487c|M.bovis_AF2122/97 QEKQKARFTYGVMEKQFRRYYEEAVRQPGKTGEELLKILESRLDNVIYRA
Rv3458c|M.tuberculosis_H37Rv QEKQKARFTYGVMEKQFRRYYEEAVRQPGKTGEELLKILESRLDNVIYRA
MLBr_01958|M.leprae_Br4923 QEKQKARFTYGVMEKQFRRYYEEAVRQPGKTGEELLKILESRLDNVIYRA
MAV_4399|M.avium_104 QEKQKARFTYGVMEKQFRRYYEEAVRQPGKTGEELLRILESRLDNVVYRA
TH_0031|M.thermoresistible__bu QEKQKARFTYGVMEKQFRRYYEEAVRRSAVTGEELLRILESRLDNVVYRA
Mflv_4971|M.gilvum_PYR-GCK QEKQKARFTYGVMEKQFRRYYEEANRLPGKTGDNLLRILESRLDNVVYRA
Mvan_1435|M.vanbaalenii_PYR-1 QEKQKARFTYGVMEKQFRRYYEEANRLPGKTGDNLLRILESRLDNVVYRA
MSMEG_1523|M.smegmatis_MC2_155 QEKQKARFSYGVMEKQFRRYYEEANRQPGKTGDNLLRILESRLDNVVYRA
MAB_3771c|M.abscessus_ATCC_199 QEKQKARFTYGVMEKQFRRYYAEAVRSSGKTGEQLLQILESRLDNVVYRA
********:************ ** * .. **::**:*********:***
MUL_0847|M.ulcerans_Agy99 GLARTRRMARQLVSHGHFSVNGVHVNVPSYRVSQYDIIDVRDNSLNTVPF
MMAR_1089|M.marinum_M GLARTRRMARQLVSHGHFSVNGVHVNVPSYRVSQYDIIDVRDNSLNTVPF
Mb3487c|M.bovis_AF2122/97 GLARTRRMARQLVSHGHFNVNGVHVNVPSYRVSQYDIVDVRDKSLNTVPF
Rv3458c|M.tuberculosis_H37Rv GLARTRRMARQLVSHGHFNVNGVHVNVPSYRVSQYDIVDVRDKSLNTVPF
MLBr_01958|M.leprae_Br4923 GLARTRRMARQLVSHGHFSVNGVHVNVPSYRVSQYDIIDIRDKSLDTVPF
MAV_4399|M.avium_104 GLARTRRMARQLVTHGHFTVNGVRVDVPSYRVSQYDIIDVREQSLNSVPF
TH_0031|M.thermoresistible__bu GLARTRRQARQLVSHGHFLVNGVKVDIPSYRVSQYDIIDIKDKSLNTVPF
Mflv_4971|M.gilvum_PYR-GCK GLARTRRMARQLVSHGHFTVNGVKVDVPSYRVSQYDIIDVKDKSINTLPF
Mvan_1435|M.vanbaalenii_PYR-1 GLARTRRMARQLVSHGHFTVNGVKVDVPSYRVSQYDIIDVKDKSINTLPF
MSMEG_1523|M.smegmatis_MC2_155 GLARTRRMARQLVSHGHFLVNGVKVDIPSYRVSQYDIIDVKEKSLNTLPF
MAB_3771c|M.abscessus_ATCC_199 GLARTRRMARQLVSHGHFTVNNVKVDVPSYRVSQYDIIDIKPKSLNTLPF
******* *****:**** **.*:*::**********:*:: :*::::**
MUL_0847|M.ulcerans_Agy99 QIARETAGDRPIPSWLQVVGGRQRILIHQLPERAQIEVPLTEQLIVEYYS
MMAR_1089|M.marinum_M QIARETAGDRPIPSWLQVVGERQRILIHQLPERAQIEVPLTEQLIVEYYS
Mb3487c|M.bovis_AF2122/97 QIARETAGERPIPSWLQVVGERQRVLIHQLPERAQIDVPLTEQLIVEYYS
Rv3458c|M.tuberculosis_H37Rv QIARETAGERPIPSWLQVVGERQRVLIHQLPERAQIDVPLTEQLIVEYYS
MLBr_01958|M.leprae_Br4923 QIARETVGDRPIPSWLQVVGEHQRILIHQLPERVQIEVPLIEQLIVEYYS
MAV_4399|M.avium_104 QIARETAGDRPVPSWLQVVGERQRILIHQLPERAQIDVPLTEQLIVEFYS
TH_0031|M.thermoresistible__bu QIARETMGDRPVPGWLQVVGERPRILVHQLPTREQIDVPLNEQLIVEFYS
Mflv_4971|M.gilvum_PYR-GCK EVARQTAGERPIPGWLQVVGERQRILVHQLPERAQIDVPLTEQLIVELYS
Mvan_1435|M.vanbaalenii_PYR-1 EVARQTAGERPIPGWLQVVGERQRILVHQLPERAQIDVPLTEQLIVELYS
MSMEG_1523|M.smegmatis_MC2_155 QIARETAGERPIPSWLQVVGERQRILVHQLPERAQIDVPLTEQLIVELYS
MAB_3771c|M.abscessus_ATCC_199 EVARETAGERPIPGWLQVVGERQRILVHQLPERAQIDVPLAEQLIVEFYS
::**:* *:**:*.****** : *:*:**** * **:*** ****** **
MUL_0847|M.ulcerans_Agy99 K
MMAR_1089|M.marinum_M K
Mb3487c|M.bovis_AF2122/97 K
Rv3458c|M.tuberculosis_H37Rv K
MLBr_01958|M.leprae_Br4923 K
MAV_4399|M.avium_104 K
TH_0031|M.thermoresistible__bu K
Mflv_4971|M.gilvum_PYR-GCK K
Mvan_1435|M.vanbaalenii_PYR-1 K
MSMEG_1523|M.smegmatis_MC2_155 K
MAB_3771c|M.abscessus_ATCC_199 K
*