For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAEQTRVHVVRHGEVHNPTGVLYGRLPGYRLSETGAAQAAAVAEYLADRDVVAVVASPLQRAQETAAPIA AAHELAIDTDPDLIESANFFEGRRVGPGDGAWRDPRVWWQLRNPFTPSWGEPYLEIAQRMATAVDKVRAR AAGHEAVCVSHQLPVWTLRLHLTGRRLWHDPRRRDCSLASVTSLVYDGDRLVDVVYSEPAGA
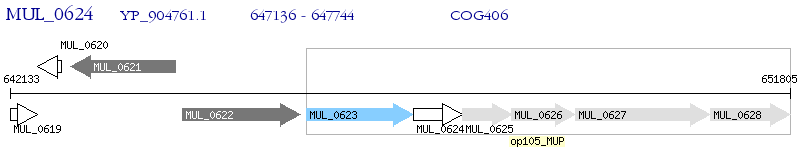
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0624 | - | - | 100% (202) | hypothetical protein MUL_0624 |
| M. ulcerans Agy99 | MUL_1324 | - | 3e-15 | 34.57% (162) | bifunctional RNase H/acid phosphatase |
| M. ulcerans Agy99 | MUL_2365 | - | 7e-07 | 40.30% (67) | hypothetical protein MUL_2365 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0538 | - | 1e-105 | 86.00% (200) | hypothetical protein Mb0538 |
| M. gilvum PYR-GCK | Mflv_0053 | - | 1e-69 | 66.00% (200) | phosphoglycerate mutase |
| M. tuberculosis H37Rv | Rv0525 | - | 1e-105 | 86.00% (200) | hypothetical protein Rv0525 |
| M. leprae Br4923 | MLBr_02413 | - | 2e-95 | 79.60% (201) | putative phosphoglycerate mutase |
| M. abscessus ATCC 19977 | MAB_3977c | - | 1e-77 | 66.67% (198) | hypothetical protein MAB_3977c |
| M. marinum M | MMAR_0871 | - | 1e-117 | 100.00% (202) | hypothetical protein MMAR_0871 |
| M. avium 104 | MAV_4620 | - | 1e-101 | 85.00% (200) | phosphoglycerate mutase family protein |
| M. smegmatis MC2 155 | MSMEG_0970 | - | 2e-70 | 65.50% (200) | phosphoglycerate mutase family protein |
| M. thermoresistible (build 8) | TH_2252 | - | 7e-84 | 72.50% (200) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_0865 | - | 8e-68 | 65.15% (198) | phosphoglycerate mutase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0624|M.ulcerans_Agy99 ----------MAEQTRVHVVRHGEVHNPTGVLYGRLPGYRLSETGAAQAA
MMAR_0871|M.marinum_M ----------MAEQTRVHVVRHGEVHNPTGVLYGRLPGYRLSETGAAQAA
Mb0538|M.bovis_AF2122/97 ----------MPEETQVHVVRHGEVHNPTGILYGRLPGFHLSATGAAQAA
Rv0525|M.tuberculosis_H37Rv ----------MPEETQVHVVRHGEVHNPTGILYGRLPGFHLSATGAAQAA
MLBr_02413|M.leprae_Br4923 ----------MAEHNRVHVVRHGEVDNPTGILYGRLAGFGLSETGRAQVA
MAV_4620|M.avium_104 ----------MGEQTRVHVVRHGEVHNPDGVLYGRLPGFHLSEAGRAQAA
MAB_3977c|M.abscessus_ATCC_199 MTDGLRASGSSQTRTIVHLMRHGEVFNPEGILYGRLPNFRLSDKGQGQAA
Mflv_0053|M.gilvum_PYR-GCK -MTDQTPGAGGANQTIVHVMRHGEVHNPEKILYGRLPDYHLSDRGQAQAR
Mvan_0865|M.vanbaalenii_PYR-1 ------------MKTVVHVMRHGEVHNPEKILYGRLADYHLSERGQAQAQ
MSMEG_0970|M.smegmatis_MC2_155 ------------MRTTVHVMRHGEVHNPAKVLYGRLPGYHLSERGRAQAR
TH_2252|M.thermoresistible__bu ----------MSERTIVHFMRHGXVHNPEGILYGRLPGYRLSERGRAQAE
.. **.:*** * ** :*****..: ** * .*.
MUL_0624|M.ulcerans_Agy99 AVAEYLADRDVVAVVASPLQRAQETAAPIAAAHELAIDTDPDLIESANFF
MMAR_0871|M.marinum_M AVAEYLADRDVVAVVASPLQRAQETAAPIAAAHELAIDTDPDLIESANFF
Mb0538|M.bovis_AF2122/97 AVADALADRDIVAVIASPLQRAQETAAPIAARHDLAVETDPDLIESANFF
Rv0525|M.tuberculosis_H37Rv AVADALADRDIVAVIASPLQRAQETAAPIAARHDLAVETDPDLIESANFF
MLBr_02413|M.leprae_Br4923 AVADALADRDVVAVIASPLQRAQETAAPIAEKHNLPVDTDPDLIESVSFF
MAV_4620|M.avium_104 AVAEALAPRDIVAVIASPLQRAQETAAPIAARHGLAVDTDPDLIESANFF
MAB_3977c|M.abscessus_ATCC_199 AVAKSLVDNKITAVVASPLLRAQQTATPIAEVHGLTIVTDDDLIEAGNSF
Mflv_0053|M.gilvum_PYR-GCK RVAEWLGSRDITHVVASPLERAQETAAPIAAAHGLAVSTDDELIESENIF
Mvan_0865|M.vanbaalenii_PYR-1 AVADWLAVRDIIHVVASPLERAQETAAPIAARHGLHINVDDELIESTNVF
MSMEG_0970|M.smegmatis_MC2_155 SAADWLAGRDIVYLVASPLERAQETAAPIAESHGLSIDTDDDLIESWNTF
TH_2252|M.thermoresistible__bu RVAEVLADRDIVLVVASPLQRAQETAAPIAARHGLTVETDDNLIESLNVF
.*. * ..: ::**** ***:**:*** * * : .* :***: . *
MUL_0624|M.ulcerans_Agy99 EGRRVGPGDGAWRDPRVWWQLRNPFTPSWGEPYLEIAQRMATAVDKVRAR
MMAR_0871|M.marinum_M EGRRVGPGDGAWRDPRVWWQLRNPFTPSWGEPYLEIAQRMATAVDKVRAR
Mb0538|M.bovis_AF2122/97 EGRRVGPGDGAWRDPRVWWQLRNPFTPSWGEPYVDIAARMTTAVDKARVR
Rv0525|M.tuberculosis_H37Rv EGRRVGPGDGAWRDPRVWWQLRNPFTPSWGEPYVDIAARMTTAVDKARVR
MLBr_02413|M.leprae_Br4923 EGRRVGLGGSTWRDPRLWWQLRNPFTPSWGESYIEIAARMTTAMDKARTR
MAV_4620|M.avium_104 EGRRISPGDGAWRDPRVWWQLRNPFRPSWGEPYTEIAARMETAVDKARAR
MAB_3977c|M.abscessus_ATCC_199 EGMKVSVGDGALRDPRNWWKLRDPFTPSWGEPYRDIASRMTAAIDRARLS
Mflv_0053|M.gilvum_PYR-GCK QGQRVSPGDGALRDPRNWWHLRNPRTPSWGEPYSHIAARMKRAVLRAREA
Mvan_0865|M.vanbaalenii_PYR-1 QGQRVSPGDGALRDPRNWWHLRNPRRPSWGEPYAEIAARMKRAVLRAREA
MSMEG_0970|M.smegmatis_MC2_155 EGQKVAPGDGALRDPRNWPRLRDPRKPSWGEPYHEVAARMMAALHRARLK
TH_2252|M.thermoresistible__bu EGQRVSPGDGALRDPRNWRFLYNPFKPSWGEPYRDIVARMTTALEKARAK
:* ::. *..: **** * * :* *****.* .:. ** *: :.*
MUL_0624|M.ulcerans_Agy99 AAGHEAVCVSHQLPVWTLRLHLTGRRLWHDPR--RRDCSLASVTSLVYDG
MMAR_0871|M.marinum_M AAGHEAVCVSHQLPVWTLRLHLTGRRLWHDPR--RRDCSLASVTSLVYDG
Mb0538|M.bovis_AF2122/97 GAGHEVVCVSHQLPVWTLRLYLTGKRLWHDPR--RRDCALASVTSLIYDG
Rv0525|M.tuberculosis_H37Rv GAGHEVVCVSHQLPVWTLRLYLTGKRLWHDPR--RRDCALASVTSLIYDG
MLBr_02413|M.leprae_Br4923 GAGHEVVCVSHQLPVWTLRLHLTGKRLWHDPR--RRDCALASVTTFVYDD
MAV_4620|M.avium_104 GTGHEVVCVSHQLPVWTLRLHLTGRRLWHDPR--KRECGLCSITTLVYDG
MAB_3977c|M.abscessus_ATCC_199 AEGAEAVCVSHQLPVWTARQHLLGKKLWHDPR--KRQCNVASLTSLVFEG
Mflv_0053|M.gilvum_PYR-GCK AHGHEAVCVSHQLPVETLRRAMTDRPLHHFPT--RRMCNLASVTSFYFHD
Mvan_0865|M.vanbaalenii_PYR-1 ADGHEAVCVSHQLPVETLRRAMTDRPLHHFPT--KRMCNLASVTSFYFHD
MSMEG_0970|M.smegmatis_MC2_155 AAGHEAVCVSHQLPVETLRRAMTGRKLAHLPLPNSRLCNLASITSFTFDD
TH_2252|M.thermoresistible__bu ATGHEAVCVSHQLPVWTLRSAMEGRRLWHDPR--KRECNLASLTSFVFDG
. * *.********* * * : .: * * * * * :.*:*:: :..
MUL_0624|M.ulcerans_Agy99 DRLVDVVYSEPAGA
MMAR_0871|M.marinum_M DRLVDVVYSEPAGA
Mb0538|M.bovis_AF2122/97 DRLVDVVYSQPAAL
Rv0525|M.tuberculosis_H37Rv DRLVDVVYSQPAAL
MLBr_02413|M.leprae_Br4923 DRLVEVEYSQPAGT
MAV_4620|M.avium_104 DRLVDVEYSEPAAP
MAB_3977c|M.abscessus_ATCC_199 EQLVDVVYSEPAG-
Mflv_0053|M.gilvum_PYR-GCK GVCVGWGYSELAGQ
Mvan_0865|M.vanbaalenii_PYR-1 DACVGWGYSELAGQ
MSMEG_0970|M.smegmatis_MC2_155 DRLIRWGYSEPWGI
TH_2252|M.thermoresistible__bu DRLVEVKYCEPAEL
: *.: