For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRLILNVIWLVFGGLWLALGYLVAALICFLLIVTIPFGFAALRIASYALWPFGRTIVEKPSAGTGALIGN VIWILLFGLWLAIGHLVSAVAMAITIIGIPLALANLKLIPVSLVPLGKEIVSADWPTRYERVAR
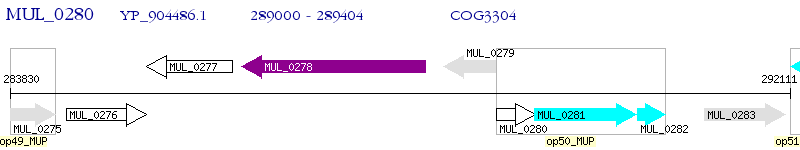
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0280 | - | - | 100% (134) | hypothetical protein MUL_0280 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0894c | - | 2e-61 | 87.90% (124) | hypothetical protein Mb0894c |
| M. gilvum PYR-GCK | Mflv_1706 | - | 3e-57 | 82.26% (124) | hypothetical protein Mflv_1706 |
| M. tuberculosis H37Rv | Rv0870c | - | 4e-62 | 88.71% (124) | hypothetical protein Rv0870c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4662 | - | 5e-73 | 100.00% (134) | hypothetical protein MMAR_4662 |
| M. avium 104 | MAV_0999 | - | 3e-59 | 81.75% (126) | hypothetical protein MAV_0999 |
| M. smegmatis MC2 155 | MSMEG_5697 | - | 6e-59 | 83.06% (124) | hypothetical protein MSMEG_5697 |
| M. thermoresistible (build 8) | TH_1070 | - | 4e-55 | 76.61% (124) | POSSIBLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_5046 | - | 2e-56 | 79.84% (124) | hypothetical protein Mvan_5046 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0280|M.ulcerans_Agy99 MRLILNVIWLVFGGLWLALGYLVAALICFLLIVTIPFGFAALRIASYALW
MMAR_4662|M.marinum_M MRLILNVIWLVFGGLWLALGYLVAALICFLLIVTIPFGFAALRIASYALW
Mb0894c|M.bovis_AF2122/97 MRLILNVIWLVFGGLWLALGYLLASLVCFLLIITIPFGFAALRIASYALW
Rv0870c|M.tuberculosis_H37Rv MRLILNVIWLVFGGLWLALGYLLASLVCFLLIITIPFGFAALRIASYALW
MAV_0999|M.avium_104 MRLILNVIWLVFGGLWMAVGYLAAALVCFLLIITIPFGFASLRIASYALW
Mflv_1706|M.gilvum_PYR-GCK MRLILNVIWLVFGGLWLALGYLLAALICFVLIVTIPFGFASLRIAAYALW
Mvan_5046|M.vanbaalenii_PYR-1 MRLALNVIWLVFGGLWLALGYLLAAIICFVLIITIPFGFASLRIAAYALW
MSMEG_5697|M.smegmatis_MC2_155 MRVLLNIIWLIFGGLWLALGYLLAALICFILIVTIPFGFASLRIASYALW
TH_1070|M.thermoresistible__bu MRLLLNVVWLIFGGLWLALGYLLASLICFLLIVTIPFGFAAFRIGMYALW
**: **::**:*****:*:*** *:::**:**:*******::**. ****
MUL_0280|M.ulcerans_Agy99 PFGRTIVEKPSAGTGALIGNVIWILLFGLWLAIGHLVSAVAMAITIIGIP
MMAR_4662|M.marinum_M PFGRTIVEKPSAGTGALIGNVIWILLFGLWLAIGHLVSAVAMAITIIGIP
Mb0894c|M.bovis_AF2122/97 PFGRTIVEKPTAGTGALISNVIWVLLFGIWLALGHLVSAAAMAVTIIGIP
Rv0870c|M.tuberculosis_H37Rv PFGRTIVEKPTAGTGALIGNVIWVLLFGIWLALGHLVSAAAMAVTIIGIP
MAV_0999|M.avium_104 PFGRTIVDKPTAGSGALIGNVIWVVLFGVWLAIGHILSAAAMALTIVGIP
Mflv_1706|M.gilvum_PYR-GCK PFGRTIVEKPGPRPGALVGNVIWVIVAGVWLAIGHITTALAMAITIVGIP
Mvan_5046|M.vanbaalenii_PYR-1 PFGRTIVEKPGPRPGALVGNVIWVIVAGVWLAIGHITTAVAMAVTIIGIP
MSMEG_5697|M.smegmatis_MC2_155 PFGRTIVEKPGPRPGALVGNVIWVILFGIWLAIGHVASAIAMAITIIGIP
TH_1070|M.thermoresistible__bu PFGRTVVDKPGPRPGALIGNIIWIVLFGIWLVLEHLITAALMAITIIGIP
*****:*:** . .***:.*:**::: *:**.: *: :* **:**:***
MUL_0280|M.ulcerans_Agy99 LALANLKLIPVSLVPLGKEIVSAD--WPTRYER-VAR-
MMAR_4662|M.marinum_M LALANLKLIPVSLVPLGKEIVSAD--WPTRYER-VAR-
Mb0894c|M.bovis_AF2122/97 LALANLKLIPVSLVPLGKDIVGVN--SQVPT-------
Rv0870c|M.tuberculosis_H37Rv LALANLKLIPVSLVPLGKDIVGVN--SQVPT-------
MAV_0999|M.avium_104 LALANLKLIPVSLMPLGKQIVPVG--SPVPHAPPVAA-
Mflv_1706|M.gilvum_PYR-GCK LALANLKLIPVSLMPLGKEIVPVD--QAHG-RVVGSVP
Mvan_5046|M.vanbaalenii_PYR-1 LALANLKMIPVSLMPLGKEIVPVD--EAGQRRIVGAVP
MSMEG_5697|M.smegmatis_MC2_155 LALANLKMIPVSLVPLGKEIVPVD--QFNNLNTERVIT
TH_1070|M.thermoresistible__bu LALANLKMIPISLMPLGKEIVPVDGLRAVPDPTGAAA-
*******:**:**:****:** ..