For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRIDRILDDARNRYPRLPADQVPDALRRGAVLVDIRPHAQRAREGEVPAALVIERNVLEWRCDRTSDARL PQAVDDDVEWVILCSEGYTSSLAAAALLDLGLHRSTDVVGGYHALVAAGVLTALSG*
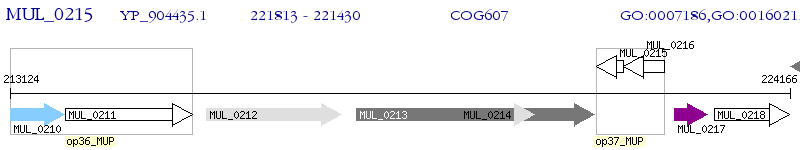
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0215 | - | - | 100% (127) | hypothetical protein MUL_0215 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1095 | - | 1e-57 | 85.04% (127) | hypothetical protein Mb1095 |
| M. gilvum PYR-GCK | Mflv_2034 | - | 4e-49 | 78.63% (117) | rhodanese domain-containing protein |
| M. tuberculosis H37Rv | Rv1066 | - | 1e-57 | 85.04% (127) | hypothetical protein Rv1066 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1183 | - | 2e-41 | 70.59% (119) | hypothetical protein MAB_1183 |
| M. marinum M | MMAR_4400 | - | 5e-67 | 97.64% (127) | hypothetical protein MMAR_4400 |
| M. avium 104 | MAV_1194 | - | 8e-55 | 79.53% (127) | hypothetical protein MAV_1194 |
| M. smegmatis MC2 155 | MSMEG_5279 | - | 1e-50 | 80.83% (120) | hypothetical protein MSMEG_5279 |
| M. thermoresistible (build 8) | TH_0676 | - | 4e-51 | 79.20% (125) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_4683 | - | 1e-47 | 79.13% (115) | rhodanese domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2034|M.gilvum_PYR-GCK MTSRIDAVLDRARARLTRLTADEVPDALARGAVLVDIRPAAQRATEGEIS
Mvan_4683|M.vanbaalenii_PYR-1 MPSRIDVVLDEARAKLTRLAADEVPAALARGAVLVDIRPAAQRAAEGEVP
MUL_0215|M.ulcerans_Agy99 MS-RIDRILDDARNRYPRLPADQVPDALRRGAVLVDIRPHAQRAREGEVP
MMAR_4400|M.marinum_M MS-RIDRILDDARNRYPRLPADQVPDALRRGAVLVDIRPQAQRAREGEVP
Mb1095|M.bovis_AF2122/97 MS-RIDRVLEAARRRYRRLAADQVPEAARRGAVLVDIRPQAQRAREGEVP
Rv1066|M.tuberculosis_H37Rv MS-RIDRVLEAARRRYRRLAADQVPEAARRGAVLVDIRPQAQRAREGEVP
MAV_1194|M.avium_104 MS-RIDVVLRSARRRFRRLPAVEVPDAVRRGAVLVDIRPQAQRFREGEVP
MSMEG_5279|M.smegmatis_MC2_155 MS-RIDTILEAARARLVRLAAEDLPAALDNGAVLVDIRPAAQRAHEGEVP
TH_0676|M.thermoresistible__bu MSSRIDIMLAAARDRLTRLPAAAVPAALRRGAVLVDIRPQAQRDREGEVP
MAB_1183|M.abscessus_ATCC_1997 MT--IHDLLASARGRLRRLPADEVPTALGRGAFLVDIRPAAQRAEEGEVI
*. *. :* ** : **.* :* * .**.****** *** ***:
Mflv_2034|M.gilvum_PYR-GCK AALIIERNVLEWRCDPTSDARIPHAVGDDVEWVVLCSEGYTSSLAAASLQ
Mvan_4683|M.vanbaalenii_PYR-1 QALVIERNVLEWRCDPTSDARLRHAVDDDVEWVVLCSQGYTSSLAAASLK
MUL_0215|M.ulcerans_Agy99 AALVIERNVLEWRCDRTSDARLPQAVDDDVEWVILCSEGYTSSLAAAALL
MMAR_4400|M.marinum_M AAVVIERNVLEWRCDPTSDARLPQAVDDDVEWVILCSEGYTSSLAAAALL
Mb1095|M.bovis_AF2122/97 GALVIERNVLEWRCDPTSDARLPQAVDDDVEWVILCSEGYTSSLAAASLL
Rv1066|M.tuberculosis_H37Rv GALVIERNVLEWRCDPTSDARLPQAVDDDVEWVILCSEGYTSSLAAASLL
MAV_1194|M.avium_104 GALVIERNVLEWRCDPTSEARLPEAVGDDVEWVIICSEGYTSSLAAAALL
MSMEG_5279|M.smegmatis_MC2_155 GALVIERNVLEWRCDPTSEAKIPQAVDDDVYWVILCSEGYTSSLAAAALL
TH_0676|M.thermoresistible__bu GALVVERNVLEWRCDPTSDARLPQAVDDDVEWVVLCSQGYTSSLAAAALQ
MAB_1183|M.abscessus_ATCC_1997 NALVIERNVLEWRLDPESDARIPQAGNHDVEWVILCQEGYTSSLAAASLQ
*:::******** * *:*:: .* ..** **::*.:*********:*
Mflv_2034|M.gilvum_PYR-GCK ELGLHRATDVIGGYHALKAVLN------------------------
Mvan_4683|M.vanbaalenii_PYR-1 DLGLRRATDVIGGYHALKSVLV------------------------
MUL_0215|M.ulcerans_Agy99 DLGLHRSTDVVGGYHALVAAGVLTALSG------------------
MMAR_4400|M.marinum_M DLGLHRSTDVVGGYHALVAAGVLTALSG------------------
Mb1095|M.bovis_AF2122/97 DLGLHRATDVVGGYRALAAGGVLAELGGAVGG--------------
Rv1066|M.tuberculosis_H37Rv DLGLHRATDVVGGYRALAAGGVLAELGGAVGG--------------
MAV_1194|M.avium_104 DIGLHRATDVIGGYHALAGAGVLSRLAGGPVGAPLANTGAGERRWI
MSMEG_5279|M.smegmatis_MC2_155 DLGLHRATDVIGGYHALVAAGQV-----------------------
TH_0676|M.thermoresistible__bu GLGLHRATDVVGGYQALAAEGVLAQLTSSG----------------
MAB_1183|M.abscessus_ATCC_1997 DIGLYRATDVIGGYKALKDKGILI----------------------
:** *:***:***:**