For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AAVGSAMRRSGSAQAAASKLFHRPPSRTAPRSNAMTAISGPTTRSLSDSTDALLRFAMRADATLTGLMGL ALAAAADPISSLTGLTSAQEYSIGAFFVLYGLLVFGLAAIPNLRRAGIGVVVGNLVYTVGAIVAAELVPM TGIGVAATVASGVFTAAFAGLQYLGVRRITA*
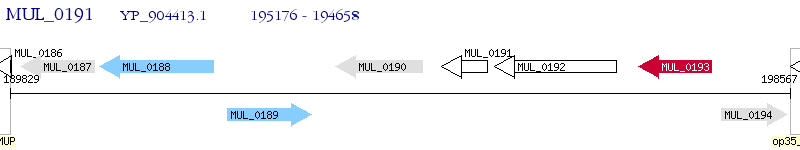
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0191 | - | - | 100% (172) | hypothetical protein MUL_0191 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2057 | - | 2e-22 | 38.46% (130) | hypothetical protein Mflv_2057 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1231 | - | 6e-13 | 36.59% (123) | hypothetical protein MAB_1231 |
| M. marinum M | MMAR_4378 | - | 6e-69 | 98.54% (137) | hypothetical protein MMAR_4378 |
| M. avium 104 | MAV_1213 | - | 8e-47 | 66.42% (137) | hypothetical protein MAV_1213 |
| M. smegmatis MC2 155 | MSMEG_5254 | - | 9e-27 | 47.45% (137) | hypothetical protein MSMEG_5254 |
| M. thermoresistible (build 8) | TH_2163 | - | 8e-30 | 54.01% (137) | conserved hypothetical protein |
| M. vanbaalenii PYR-1 | Mvan_4659 | - | 3e-26 | 46.03% (126) | hypothetical protein Mvan_4659 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0191|M.ulcerans_Agy99 MAAVGSAMRRSGSAQAAASKLFHRPPSRTAPRSNAMTAISGPTTRSLSDS
MMAR_4378|M.marinum_M -----------------------------------MTAISGPTTRPLSDS
MAV_1213|M.avium_104 --------------------------------MSTVSGVSGVTVRPLYDS
MSMEG_5254|M.smegmatis_MC2_155 --------------------------------------MTAITTRPLSDS
TH_2163|M.thermoresistible__bu --------------------------VVFDTHTAEEAVMTAITTRPLSDS
Mflv_2057|M.gilvum_PYR-GCK --------------------------------------MTATTV-PGADT
Mvan_4659|M.vanbaalenii_PYR-1 --------------------------------------MATTISARLNES
MAB_1231|M.abscessus_ATCC_1997 ---------------------------------MSIASQNASVLRTRFAQ
MUL_0191|M.ulcerans_Agy99 TDALLRFAMRADATLTGLMGLALAAAADPISSLTGLTSAQEYSIGAFFVL
MMAR_4378|M.marinum_M TDALLRFVMRADATLTGLMGLALAAAADPISSLTGLTSAQEYSIGAFFVL
MAV_1213|M.avium_104 TDSLLRFAVRADATLTGLCGLAVAVAADPLSSLTGLTSFQEYIGGAFFVL
MSMEG_5254|M.smegmatis_MC2_155 TDSLLRFALRADATVCAGVGLFTAMLADPLARVAGLSATAGWVIGAALVG
TH_2163|M.thermoresistible__bu TDSLLRFAMRADATVCGGLGLFIAFAADPLSGLTGLTTAGQFLAGAALAG
Mflv_2057|M.gilvum_PYR-GCK RSGLLRFAMRLDAMLVGITGIPFVVAAGWLSSLTGIPVAVEYALGVFFLA
Mvan_4659|M.vanbaalenii_PYR-1 TDTLLRVAMRADAVLSGLTGIGLLIFAPQVAEISGTTPSVEYTIGASFVV
MAB_1231|M.abscessus_ATCC_1997 PDALLRFGIGLDGVATGTAAVGLLIAAKWLVEPLGPSLQFQVGHAAALIG
. ***. : *. . .: * : * . .. :
MUL_0191|M.ulcerans_Agy99 YGLLVFGLAAIPNLRR--AGIGVVVGNLVYTVGAI--VAAELVPMTGIGV
MMAR_4378|M.marinum_M YGLLVFGLAAIPNLRR--AGIGVVVGNLVYTVGAI--VAAELVPMTGIGV
MAV_1213|M.avium_104 YGLVVFSLGALPDLRR--AGISIVAANAVFTLAAI--VAAGVLPLTGAGV
MSMEG_5254|M.smegmatis_MC2_155 YGALLYVLAALPAIRK--VGVAVATGNTVFAAASVTAVLAGWLPLTGAGT
TH_2163|M.thermoresistible__bu YGFLLYVLAAAGALRR--IGIGVVAANTAFVATALTVLAAGWLHLTAIGN
Mflv_2057|M.gilvum_PYR-GCK YGGVVYWLAGLDDVRP--GALATIAMNALFTIGFVGAEVAGVWPLTGWGI
Mvan_4659|M.vanbaalenii_PYR-1 FSAVVLALAARPAVRG--SGLVLALGNALFTVATIAVVVIGVWPLTTVGI
MAB_1231|M.abscessus_ATCC_1997 YGVLAFALSRADRSRLGTIGVVYIAANLLATVLYVMAGVMTWVPLTTAGV
:. : *. * .: * . : :* *
MUL_0191|M.ulcerans_Agy99 AATVASGVFTAAFAGLQYLGVRRITA-
MMAR_4378|M.marinum_M AATVASGVFTAAFAGLQYLGVRRITA-
MAV_1213|M.avium_104 ALTLASAAYTAAFAAVQYAGVRRLETA
MSMEG_5254|M.smegmatis_MC2_155 ELLLGFVAATAALAWLQYRGVRRLA--
TH_2163|M.thermoresistible__bu TAALAFTALTLALGYLQYLGVRRLA--
Mflv_2057|M.gilvum_PYR-GCK GVLLGGALYTAVIGAVQYVGLRRLR--
Mvan_4659|M.vanbaalenii_PYR-1 ALTLGTGVYTLVMAELQYQGVRRIPK-
MAB_1231|M.abscessus_ATCC_1997 TLCIAFGVYTAVMADIQFLGLRRLRSA
:. * .:. :*: *:**: