For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTQLQPPVTVTPPLPTRRNAELLLLCFAAVITIAALFIVEANQDRAIHWDLASYGLAFLTLFGFAHLAIR RFAPFTDPLLLPVVALLNGLGLVMIHRLDLVDKQIGHRAHPSANQQMLWTLVGVASFALVVTFLKDHRQL ARYGYTCGLAGLVLLAIPAVLPAALSEENGSKIWIRLPGFSIQPAEFSKILLLIFFSAVLVAKRGLFTSA GKHLMGMTLPRPRDLAPLLAAWVASVGVMVFEKDLGTSLLLYASFLVVLYLATQRFSWVIIGLTLFTAGS VVAYFVFHHVRVRVQNWLDPFADPDGTGYQMVQALFGFATGGIFGTGLGNGQPDTVPAASTDFIIAAFGE ELGLVGLAAILMLYTIVIIRGLRTAIATRDSFGKLLAAGLASTLAIQLFIVVGGVTKLIPLTGLTTPWMS YGGSSLLANYVLLAILARISHGARRPLRTRPRNTSSIAAAGTEVIERV*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
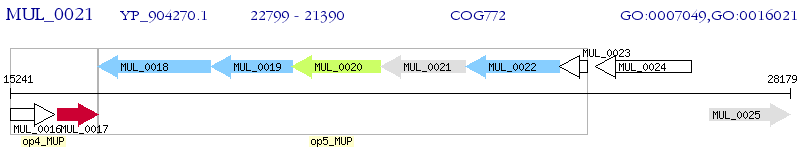
| M. ulcerans Agy99 | MUL_0021 | rodA | - | 100% (469) | cell division protein RodA |
| M. ulcerans Agy99 | MUL_3502 | ftsW | 5e-42 | 32.27% (406) | FtsW-like protein FtsW |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0017c | rodA | 0.0 | 89.55% (469) | cell division protein RodA |
| M. gilvum PYR-GCK | Mflv_0809 | - | 0.0 | 77.23% (470) | cell cycle protein |
| M. tuberculosis H37Rv | Rv0017c | rodA | 0.0 | 89.55% (469) | cell division protein RodA |
| M. leprae Br4923 | MLBr_00019 | rodA | 0.0 | 82.94% (469) | putative cell-division protein |
| M. abscessus ATCC 19977 | MAB_0036c | - | 0.0 | 69.81% (467) | cell division protein RodA |
| M. marinum M | MMAR_0019 | rodA | 0.0 | 99.79% (469) | cell division protein RodA |
| M. avium 104 | MAV_0021 | - | 0.0 | 85.29% (469) | cell cycle protein, FtsW/RodA/SpoVE family protein |
| M. smegmatis MC2 155 | MSMEG_0032 | - | 0.0 | 78.77% (471) | cell cycle protein, FtsW/RodA/SpoVE family protein |
| M. thermoresistible (build 8) | TH_0841 | rodA | 0.0 | 78.40% (463) | PROBABLE CELL DIVISION PROTEIN RODA |
| M. vanbaalenii PYR-1 | Mvan_0026 | - | 0.0 | 77.02% (470) | cell cycle protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0809|M.gilvum_PYR-GCK MTTQPQ----------APVAVTPALPNRRNAELLLLGFAAFLTTIALLLV
Mvan_0026|M.vanbaalenii_PYR-1 MTTQPQ----------PAVAVTPPLPNRRNAELFLLGFAALITTVALLLV
MSMEG_0032|M.smegmatis_MC2_155 MTTQPQ----------SPVTVTPPLPNRRNAELLLLGFAALITTVALLIV
TH_0841|M.thermoresistible__bu ------------------VTVVPPLPTRRNAELLLLAFAAVITMVALLLV
MUL_0021|M.ulcerans_Agy99 MTTQLQ----------PPVTVTPPLPTRRNAELLLLCFAAVITIAALFIV
MMAR_0019|M.marinum_M MTTQLQ----------PPVTVTPPLPTRRNAELLLLCFAAVITIAALFIV
Mb0017c|M.bovis_AF2122/97 MTTRLQ----------APVAVTPPLPTRRNAELLLLCFAAVITFAALLVV
Rv0017c|M.tuberculosis_H37Rv MTTRLQ----------APVAVTPPLPTRRNAELLLLCFAAVITFAALLVV
MLBr_00019|M.leprae_Br4923 MTTQLQ----------PVVTVTPPLPTRRNAELLLLGFAAVITVAALAIV
MAV_0021|M.avium_104 MTTQLQ----------PPVAVTPPLPTRRNAELLLLGFAATITVAALLIV
MAB_0036c|M.abscessus_ATCC_199 MTTAPQTAVGPSGRGSGAVSAPPTQSTRRSAELGLLGFAAAITTVALVLV
*:. *. ..**.*** ** *** :* ** :*
Mflv_0809|M.gilvum_PYR-GCK EANQEQGLHWDLAQYTVAYLALFTGAHLAVRRFAPYADPLLLPVVALLNG
Mvan_0026|M.vanbaalenii_PYR-1 EANQEQGLHWDLAQYTVAYLALFTGAHLAVRRFAPYADPLLLPVVALLNG
MSMEG_0032|M.smegmatis_MC2_155 EANQEQGVTWDLAHYMVGYLALFGAAHLAVRRFAPYADPLLLPVVAVLNG
TH_0841|M.thermoresistible__bu EANQERGVSWHLIQYGAAYLALFGGAHLAVRRFAPYADPLLLPVVALLNG
MUL_0021|M.ulcerans_Agy99 EANQDRAIHWDLASYGLAFLTLFGFAHLAIRRFAPFTDPLLLPVVALLNG
MMAR_0019|M.marinum_M EANQDRAIHWDLASYGLAFLTLFGFAHLAIRRFAPFTDPLLLPVVALLNG
Mb0017c|M.bovis_AF2122/97 QANQDQGVPWDLTSYGLAFLTLFGSAHLAIRRFAPYTDPLLLPVVALLNG
Rv0017c|M.tuberculosis_H37Rv QANQDQGVPWDLTSYGLAFLTLFGSAHLAIRRFAPYTDPLLLPVVALLNG
MLBr_00019|M.leprae_Br4923 EANQERNFRWYLAGYGLIFWSLFASAHLAIRRFAPYTDPLLLPIVALLNG
MAV_0021|M.avium_104 EANQQHDLRWNAVSYGLVFLVVFGSAHVAIRRFAPYTDPLLLPIVALLNG
MAB_0036c|M.abscessus_ATCC_199 EANLERGLSWDLLGYGFAYLVLFVLAHMAVRRWAPYADPLLLPCVALLNG
:** :: . * * : :* **:*:**:**::****** **:***
Mflv_0809|M.gilvum_PYR-GCK LGLVMIHRLDLAQ--GENTSQGLGG-SANQQMLWTLVGVLGFSAVVIFLR
Mvan_0026|M.vanbaalenii_PYR-1 LGLVMIHRLDLAE--GETTAQGLGG-TANQQMLWTLVGVLGFSAVVIFLR
MSMEG_0032|M.smegmatis_MC2_155 LGLVMIHRLDLAE--GELTADGLGG-SANQQMLWTLVGVVAFALVVVFLK
TH_0841|M.thermoresistible__bu LGLVMIHRLDLAA--GELSGQAVESPSATQQTLWTLAAVIGFSLVVVFLK
MUL_0021|M.ulcerans_Agy99 LGLVMIHRLDLVD--KQIGHRAHPS--ANQQMLWTLVGVASFALVVTFLK
MMAR_0019|M.marinum_M LGLVMIHRLDLVD--KQIGHRAHPS--ANQQMLWTLVGVASFALVVTFLK
Mb0017c|M.bovis_AF2122/97 LGLVMIHRLDLVD--NEIGEHRHPS--ANQQMLWTLVGVAAFALVVTFLK
Rv0017c|M.tuberculosis_H37Rv LGLVMIHRLDLVD--NEIGEHRHPS--ANQQMLWTLVGVAAFALVVTFLK
MLBr_00019|M.leprae_Br4923 LGLVMIHRLDLVD--NDVTGHHHTS--AAQQMLWTLVGVAAFVLVMTVLK
MAV_0021|M.avium_104 LGLVMIHRLDLVT--NQLSGRHHPS--ATQQMLWTLIGVVTFALVVTFLK
MAB_0036c|M.abscessus_ATCC_199 LGLVMIHRLDLVHSYNDGDADQVGDNNATQQMLWTTVGLIALAAVLATLK
***********. : . * ** *** .: : *: *:
Mflv_0809|M.gilvum_PYR-GCK DHRMLSRYGYVCGLTGLILLAIPAVLPRSMSEQNGAKIWIQFHGFSIQPA
Mvan_0026|M.vanbaalenii_PYR-1 DHRTLSRYGYVCGLTGLVLLAIPAILPRSMSEQNGAKIWIQFEGFSIQPA
MSMEG_0032|M.smegmatis_MC2_155 DHRMLSRYGYVFGLTGLVLLVIPALLPAKYSEQYGAKIWIQFPGFSIQPA
TH_0841|M.thermoresistible__bu DHRLLSRYGYLCGATGLILLAIPAVLPRRFSEQGGAKIWIQFPGFSIQPA
MUL_0021|M.ulcerans_Agy99 DHRQLARYGYTCGLAGLVLLAIPAVLPAALSEENGSKIWIRLPGFSIQPA
MMAR_0019|M.marinum_M DHRQLARYGYTCGLAGLVLLAIPAVLPAALSEENGSKIWIRLPGFSIQPA
Mb0017c|M.bovis_AF2122/97 DHRQLARYGYICGLAGLVFLAVPALLPAALSEQNGAKIWIRLPGFSIQPA
Rv0017c|M.tuberculosis_H37Rv DHRQLARYGYICGLAGLVFLAVPALLPAALSEQNGAKIWIRLPGFSIQPA
MLBr_00019|M.leprae_Br4923 DHRQLARYGYISGLTGLVFLAIPAPLP----EQNGAKIWIRFPGFSIQPA
MAV_0021|M.avium_104 DHRQLARYGYICGLVGLVFLVIPALLPASLSEQNGAKIWIRFPGFSIQPA
MAB_0036c|M.abscessus_ATCC_199 DHRVLARYGYTFGIVGCVLLAIPALLPTSMSEQYGAKIWIELPFLSIQPA
*** *:**** * .* ::*.:** ** *: *:****.: :*****
Mflv_0809|M.gilvum_PYR-GCK EFSKILLLVFFAAVLVDKRSLFTSAGTHVLGMDLPRPRDLAPLLAAWIAS
Mvan_0026|M.vanbaalenii_PYR-1 EFSKILLLIFFAAVLVSKRDLFTSAGKHVLGMDLPRPRDLAPLLAAWIAS
MSMEG_0032|M.smegmatis_MC2_155 EFSKILLLIFFSAVLVSKRSLFTSAGKHVLGVDLPRPRDLAPLLVAWAAS
TH_0841|M.thermoresistible__bu EFSKILLLVFFAAVLVEKRSLFTSAGKHFLGMDLPRPRDLAPLLAAWILS
MUL_0021|M.ulcerans_Agy99 EFSKILLLIFFSAVLVAKRGLFTSAGKHLMGMTLPRPRDLAPLLAAWVAS
MMAR_0019|M.marinum_M EFSKILLLIFFSAVLVAKRGLFTSAGKHLMGMTLPRPRDLAPLLAAWVAS
Mb0017c|M.bovis_AF2122/97 EFSKILLLIFFSAVLVAKRGLFTSAGKHLLGMTLPRPRDLAPLLAAWVIS
Rv0017c|M.tuberculosis_H37Rv EFSKILLLIFFSAVLVAKRGLFTSAGKHLLGMTLPRPRDLAPLLAAWVIS
MLBr_00019|M.leprae_Br4923 EFSKILLLIFFAAVLVAKRSLFTSAGKHLIGMTLPRPRDLAPLLAAWVIS
MAV_0021|M.avium_104 EFSKILLLIFFSAVLIAKRGLFTSVGKHFMGLTLPRPRDLAPLLAAWVIS
MAB_0036c|M.abscessus_ATCC_199 EAAKILLLIFFAAFLMTKRTLFRTAGRNYLGIELPRPRDLGPLLAIWGIS
* :*****:**:*.*: ** ** :.* : :*: *******.***. * *
Mflv_0809|M.gilvum_PYR-GCK IGVMIFEKDLGTSLLLYASFLVMVYIATERFSWVVLGLALFAAGSIAAYF
Mvan_0026|M.vanbaalenii_PYR-1 IAVMIFEKDLGTSLLLYASFLVMVYIATERFSWVVLGLALFAAGSVAAYY
MSMEG_0032|M.smegmatis_MC2_155 VGIMIFEKDLGTSLLLYASFLVLLYVATERISWVVIGLALFAAGSVVAYN
TH_0841|M.thermoresistible__bu VGVMVFEKDLGTSLLLYTSFLVLVYIATERLSWVVIGLALFAAGSFVAYH
MUL_0021|M.ulcerans_Agy99 VGVMVFEKDLGTSLLLYASFLVVLYLATQRFSWVIIGLTLFTAGSVVAYF
MMAR_0019|M.marinum_M VGVMVFEKDLGTSLLLYASFLVVLYLATQRFSWVIIGLTLFTAGSVVAYF
Mb0017c|M.bovis_AF2122/97 VGVMVFEKDLGASLLLYTSFLVVVYLATQRFSWVVIGLTLFAAGTLVAYF
Rv0017c|M.tuberculosis_H37Rv VGVMVFEKDLGASLLLYTSFLVVVYLATQRFSWVVIGLTLFAAGTLVAYF
MLBr_00019|M.leprae_Br4923 VSVMVFEKDLGTSLLLYASFLVVVYLATQRLSWVIIGLVLFTAGSTIAYF
MAV_0021|M.avium_104 VGVMAFEKDLGTSLLLYTSFLVVVYLATQRFSWVAIGLVLFVAGSVVAYY
MAB_0036c|M.abscessus_ATCC_199 VGVMVFEKDLGASLLFYSSFLIVLYVATARISWVLIGLALFAAGSVIAYF
:.:* ******:***:*:***:::*:** *:*** :**.**.**: **
Mflv_0809|M.gilvum_PYR-GCK LFDHVRVRVQTWRDPFADPDGAGYQMVQSLFSFATGGIFGTGLGNGQPGT
Mvan_0026|M.vanbaalenii_PYR-1 LFDHVRVRVQTWRDPFADPDGAGYQMVQSLFSFATGGIFGTGLGNGQPGT
MSMEG_0032|M.smegmatis_MC2_155 IFDHVQVRVQTWLDPFADPEGAGYQMVQSLFSFATGGIFGTGLGNGQPGT
TH_0841|M.thermoresistible__bu IFDHVRLRVQTWLDPFADFDGGGYQMAQSLFSFATGGIFGTGLGNGQPGT
MUL_0021|M.ulcerans_Agy99 VFHHVRVRVQNWLDPFADPDGTGYQMVQALFGFATGGIFGTGLGNGQPDT
MMAR_0019|M.marinum_M VFHHVRVRVQNWLDPFADPDGTGYQMVQALFSFATGGIFGTGLGNGQPDT
Mb0017c|M.bovis_AF2122/97 IFEHVRLRVQTWLDPFADPDGTGYQIVQSLFSFATGGIFGTGLGNGQPDT
Rv0017c|M.tuberculosis_H37Rv IFEHVRLRVQTWLDPFADPDGTGYQIVQSLFSFATGGIFGTGLGNGQPDT
MLBr_00019|M.leprae_Br4923 TFEHIRVRMQVWWDPFTNLDVGGYQIVQSLFSFATGGIFGTGLGNGQPDA
MAV_0021|M.avium_104 IFAHVRVRVQMWWDPFSDPDGSGYQIVQSLFSFATGGIFGTGLGNGQPDT
MAB_0036c|M.abscessus_ATCC_199 LFGHVQVRVQNWWDPFTDPDGAGYQMVQSLFSFATGGIFGTGLGNGQPGT
* *:::*:* * ***:: : ***:.*:**.****************.:
Mflv_0809|M.gilvum_PYR-GCK VPAASTDFIIAAIGEELGLVGLSAVLMLYTILIIRGLRTAIAIRDSFGKL
Mvan_0026|M.vanbaalenii_PYR-1 VPAASTDFIVAAIGEELGLVGLSAVLMLYTIVIIRGLRTAIAVRDSFGKL
MSMEG_0032|M.smegmatis_MC2_155 VPAAATDFITAAIGEELGLVGLAGVLMLYTILIIRGLRTAIAVRDSFGKL
TH_0841|M.thermoresistible__bu VPAASTDFIIAAVGEELGLVGLAAVLMLYTIVIIRGLRTAIAVRDSFGKL
MUL_0021|M.ulcerans_Agy99 VPAASTDFIIAAFGEELGLVGLAAILMLYTIVIIRGLRTAIATRDSFGKL
MMAR_0019|M.marinum_M VPAASTDFIIAAFGEELGLVGLAAILMLYTIVIIRGLRTAIATRDSFGKL
Mb0017c|M.bovis_AF2122/97 VPAASTDFIIAAFGEELGLVGLTAILMLYTIVIIRGLRTAIATRDSFGKL
Rv0017c|M.tuberculosis_H37Rv VPAASTDFIIAAFGEELGLVGLTAILMLYTIVIIRGLRTAIATRDSFGKL
MLBr_00019|M.leprae_Br4923 IPAASTDFIIAVFGEELGLVGLAALLMLYTIVIVRGLRTAIATRDSFGKL
MAV_0021|M.avium_104 VPAASTDFIIAAFGEELGLVGLAAILMLYTIVIVRGLRTAIATRDSFGKL
MAB_0036c|M.abscessus_ATCC_199 VPAASTDFIIAAVGEELGLVGLAGVLMLYTIVIIRGFRTALAVRDSFGKL
:***:**** *..*********:.:******:*:**:***:* *******
Mflv_0809|M.gilvum_PYR-GCK LAAGLAATLAIQLFIVVGGVTKLIPLTGLTTPWMSYGGSSLVANYLLLAI
Mvan_0026|M.vanbaalenii_PYR-1 LAAGLSATLAIQLFIVVGGVTKLIPLTGLTTPWMSYGGSSLVANYLLLAI
MSMEG_0032|M.smegmatis_MC2_155 LAAGLASTLAIQLFIVVGGVTRLIPLTGLTTPWMSYGGSSLLANYVLLAI
TH_0841|M.thermoresistible__bu LAAGLASTLALQLFIVVGGVTKLIPLTGLTTPWMSYGGSSLLANYLLLAI
MUL_0021|M.ulcerans_Agy99 LAAGLASTLAIQLFIVVGGVTKLIPLTGLTTPWMSYGGSSLLANYVLLAI
MMAR_0019|M.marinum_M LAAGLASTLAIQLFIVVGGVTKLIPLTGLTTPWMSYGGSSLLANYVLLAI
Mb0017c|M.bovis_AF2122/97 LAAGLSSTLAIQLFIVVGGVTRLIPLTGLTTPWMSYGGSSLLANYILLAI
Rv0017c|M.tuberculosis_H37Rv LAAGLSSTLAIQLFIVVGGVTRLIPLTGLTTPWMSYGGSSLLANYILLAI
MLBr_00019|M.leprae_Br4923 LAAGLASTLAIQLFIVSGGVTTLIPLTGLTTPWMSYGGSSLLANYVLLAI
MAV_0021|M.avium_104 LAAGLASTLAIQLFIVVGGVTQLIPLTGLTTPWMSYGGSSLLANYVLLAI
MAB_0036c|M.abscessus_ATCC_199 LAVGLSAALAMQLFIVVGGVTKLIPQTGLTTPWMSYGGSSLIANYVLLAI
**.**:::**:***** **** *** ***************:***:****
Mflv_0809|M.gilvum_PYR-GCK LVRISHNARKPIQTRPLPPGS--IAGASTEVIQKV
Mvan_0026|M.vanbaalenii_PYR-1 LVRISHSARRPIVTRSLPPGN--IAGASTEVIQKV
MSMEG_0032|M.smegmatis_MC2_155 LVVISHAARRPITTSRPPNATP-IAAASTEVIEKV
TH_0841|M.thermoresistible__bu LLRISHAARSPLDTRKTNPTP--IATAKTEVIEKV
MUL_0021|M.ulcerans_Agy99 LARISHGARRPLRTRPRNTSS--IAAAGTEVIERV
MMAR_0019|M.marinum_M LARISHGARRPLRTRPRNTSS--IAAAGTEVIERV
Mb0017c|M.bovis_AF2122/97 LARISHGARRPLRTRPRNKSP--ITAAGTEVIERV
Rv0017c|M.tuberculosis_H37Rv LARISHGARRPLRTRPRNKSP--ITAAGTEVIERV
MLBr_00019|M.leprae_Br4923 LARISHSARHPLRSRPHNTSP--IAVASTEVIERV
MAV_0021|M.avium_104 LARISHSARRPLRTRARTEPS--IAAASTEVIEKV
MAB_0036c|M.abscessus_ATCC_199 LLRISHIARRPLSPHASSSPAAPLAEARTELIDRV
* *** ** *: . :: * **:*::*