For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSEDSATVAVTDDSFSTDVLGSSKPVLVDFWATWCGPCKMVAPVLEEIAAEKGDQLTVAKIDVDANPATA RDFQVVSIPTMILFKDGAPVKRIVGAKGKAALLRELSDAL
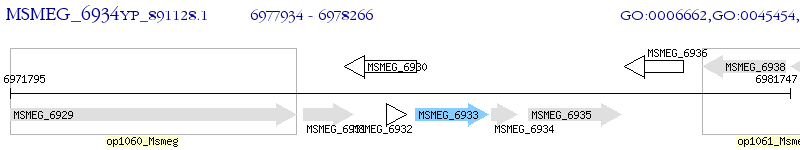
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6934 | trx | - | 100% (110) | thioredoxin |
| M. smegmatis MC2 155 | MSMEG_3138 | trx | 3e-15 | 42.27% (97) | thioredoxin |
| M. smegmatis MC2 155 | MSMEG_5786 | - | 1e-09 | 34.57% (81) | thiredoxin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3945 | trxC | 8e-48 | 81.65% (109) | thioredoxin trxC (TRX) (MPT46) |
| M. gilvum PYR-GCK | Mflv_0838 | - | 2e-48 | 84.11% (107) | thioredoxin |
| M. tuberculosis H37Rv | Rv3914 | trxC | 8e-48 | 81.65% (109) | thioredoxin trxC (TRX) (MPT46) |
| M. leprae Br4923 | MLBr_02703 | trx | 4e-44 | 78.85% (104) | bifunctional thioredoxin reductase/thioredoxin |
| M. abscessus ATCC 19977 | MAB_4941 | - | 7e-42 | 75.73% (103) | thioredoxin (Trx) |
| M. marinum M | MMAR_5478 | trxC | 2e-48 | 83.33% (108) | thioredoxin TrxC |
| M. avium 104 | MAV_5302 | trx | 2e-48 | 84.76% (105) | thioredoxin |
| M. thermoresistible (build 8) | TH_2412 | trxC | 2e-44 | 80.19% (106) | THIOREDOXIN TRXC (TRX) (MPT46) |
| M. ulcerans Agy99 | MUL_5067 | trxC | 2e-48 | 83.33% (108) | thioredoxin TrxC |
| M. vanbaalenii PYR-1 | Mvan_6068 | - | 2e-48 | 83.64% (110) | thioredoxin |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6934|M.smegmatis_MC2_155 --------------------------------------------------
TH_2412|M.thermoresistible__bu --------------------------------------------------
Mflv_0838|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_6068|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5478|M.marinum_M --------------------------------------------------
MUL_5067|M.ulcerans_Agy99 --------------------------------------------------
MAV_5302|M.avium_104 --------------------------------------------------
Mb3945|M.bovis_AF2122/97 --------------------------------------------------
Rv3914|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02703|M.leprae_Br4923 MNTTPSAHETIHEVIVIGSGPAGYTAALYAARAQLTPLVFEGTSFGGALM
MAB_4941|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6934|M.smegmatis_MC2_155 --------------------------------------------------
TH_2412|M.thermoresistible__bu --------------------------------------------------
Mflv_0838|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_6068|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5478|M.marinum_M --------------------------------------------------
MUL_5067|M.ulcerans_Agy99 --------------------------------------------------
MAV_5302|M.avium_104 --------------------------------------------------
Mb3945|M.bovis_AF2122/97 --------------------------------------------------
Rv3914|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02703|M.leprae_Br4923 TTTEVENYPGFRNGITGPELMDDMREQALRFGAELRTEDVESVSLRGPIK
MAB_4941|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6934|M.smegmatis_MC2_155 --------------------------------------------------
TH_2412|M.thermoresistible__bu --------------------------------------------------
Mflv_0838|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_6068|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5478|M.marinum_M --------------------------------------------------
MUL_5067|M.ulcerans_Agy99 --------------------------------------------------
MAV_5302|M.avium_104 --------------------------------------------------
Mb3945|M.bovis_AF2122/97 --------------------------------------------------
Rv3914|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02703|M.leprae_Br4923 SVVTAEGQTYQARAVILAMGTSVRYLQIPGEQELLGRGVSACATCDGSFF
MAB_4941|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6934|M.smegmatis_MC2_155 --------------------------------------------------
TH_2412|M.thermoresistible__bu --------------------------------------------------
Mflv_0838|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_6068|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5478|M.marinum_M --------------------------------------------------
MUL_5067|M.ulcerans_Agy99 --------------------------------------------------
MAV_5302|M.avium_104 --------------------------------------------------
Mb3945|M.bovis_AF2122/97 --------------------------------------------------
Rv3914|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02703|M.leprae_Br4923 RGQDIAVIGGGDSAMEEALFLTRFARSVTLVHRRDEFRASKIMLGRARNN
MAB_4941|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6934|M.smegmatis_MC2_155 --------------------------------------------------
TH_2412|M.thermoresistible__bu --------------------------------------------------
Mflv_0838|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_6068|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5478|M.marinum_M --------------------------------------------------
MUL_5067|M.ulcerans_Agy99 --------------------------------------------------
MAV_5302|M.avium_104 --------------------------------------------------
Mb3945|M.bovis_AF2122/97 --------------------------------------------------
Rv3914|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02703|M.leprae_Br4923 DKIKFITNHTVVAVNGYTTVTGLRLRNTTTGEETTLVVTGVFVAIGHEPR
MAB_4941|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6934|M.smegmatis_MC2_155 --------------------------------------------------
TH_2412|M.thermoresistible__bu --------------------------------------------------
Mflv_0838|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_6068|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5478|M.marinum_M --------------------------------------------------
MUL_5067|M.ulcerans_Agy99 --------------------------------------------------
MAV_5302|M.avium_104 --------------------------------------------------
Mb3945|M.bovis_AF2122/97 --------------------------------------------------
Rv3914|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02703|M.leprae_Br4923 SSLVSDVVDIDPDGYVLVKGRTTSTSMDGVFAAGDLVDRTYRQAITAAGS
MAB_4941|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6934|M.smegmatis_MC2_155 -----------------------------------------MSE---DSA
TH_2412|M.thermoresistible__bu -----------------------------------------MS------G
Mflv_0838|M.gilvum_PYR-GCK -----------------------------------------MSD---S-S
Mvan_6068|M.vanbaalenii_PYR-1 -----------------------------------------MSD---SKN
MMAR_5478|M.marinum_M -----------------------------------------MTDPEKSSA
MUL_5067|M.ulcerans_Agy99 -----------------------------------------MIDSEKSSA
MAV_5302|M.avium_104 -----------------------------------------MTDAEKAIA
Mb3945|M.bovis_AF2122/97 -----------------------------------------MTDSEKS-A
Rv3914|M.tuberculosis_H37Rv -----------------------------------------MTDSEKS-A
MLBr_02703|M.leprae_Br4923 GCAAAIDAERWLAEHAGSKANETTEETGDVDSTDTTDWSTAMTDAKNAGV
MAB_4941|M.abscessus_ATCC_1997 -----------------------------------------MSD----HA
*
MSMEG_6934|M.smegmatis_MC2_155 TVAVTDDSFSTDVLGSSKPVLVDFWATWCGPCKMVAPVLEEIAAEKGDQL
TH_2412|M.thermoresistible__bu TVTVTDSTFKTDVLDSDTPVLVDFWADWCGPCKMVAPVLEEIANEKSGTL
Mflv_0838|M.gilvum_PYR-GCK TVTVTDDSFSDDVLSSGTPVLVDFWATWCGPCKMVAPVLEEIASEKAGSL
Mvan_6068|M.vanbaalenii_PYR-1 TVTVTDDSFSDDVLSSGTPVLVDFWATWCGPCKMVAPVLEEIASEKAGSL
MMAR_5478|M.marinum_M TIEVSDASFSTEVLSSNKPVLVDFWATWCGPCKMVAPVLEEIATERADSL
MUL_5067|M.ulcerans_Agy99 TIEVSDASFSTEVLSSNKPVLVDFWATWCGPCKMVAPVLEEIATERADNL
MAV_5302|M.avium_104 TIEVSDASFSTDVLASNKPVLVDFWATWCGPCKMVAPVLEEIASERGDHL
Mb3945|M.bovis_AF2122/97 TIKVTDASFATDVLSSNKPVLVDFWATWCGPCKMVAPVLEEIATERATDL
Rv3914|M.tuberculosis_H37Rv TIKVTDASFATDVLSSNKPVLVDFWATWCGPCKMVAPVLEEIATERATDL
MLBr_02703|M.leprae_Br4923 TIEVTDASFFADVLSSNKPVLVDFWATWCGPCKMVAPVLEEIASEQRNQL
MAB_4941|M.abscessus_ATCC_1997 TVTVTDDSFQEDVVSSNKPVLVDFWATWCGPCKMVAPVLEEIAKDHGEAL
*: *:* :* :*: *..******** **************** :: *
MSMEG_6934|M.smegmatis_MC2_155 TVAKIDVDANPATARDFQVVSIPTMILFKDGAPVKRIVGAKGKAALLREL
TH_2412|M.thermoresistible__bu KVAKLDVDANPEAARDFQVVSIPTMILFKGGTPVKRIVGAKGKAALLREI
Mflv_0838|M.gilvum_PYR-GCK TVAKLDVDENPATARDFQVVSIPTMILFKDGQPVKRIVGAKGKAALLREI
Mvan_6068|M.vanbaalenii_PYR-1 TVAKIDVDENPATARDFQVVSIPTMILFKDGKPVKRIVGAKGKAALLREL
MMAR_5478|M.marinum_M TVAKLDVDANPETARNFQVVSIPTMILFKDGEPVKRIVGAKGKAALLREL
MUL_5067|M.ulcerans_Agy99 TVAKLDVDANPETARNFQVVSIPTMILFKDGEPVKRIVGAKGKAALLREL
MAV_5302|M.avium_104 TVAKLDVDANPETAQNFQVVSIPTLILFKDGQPVKRIVGAKGKAALLREL
Mb3945|M.bovis_AF2122/97 TVAKLDVDTNPETARNFQVVSIPTLILFKDGQPVKRIVGAKGKAALLREL
Rv3914|M.tuberculosis_H37Rv TVAKLDVDTNPETARNFQVVSIPTLILFKDGQPVKRIVGAKGKAALLREL
MLBr_02703|M.leprae_Br4923 TVAKLDVDTNPEMAREFQVVSIPTMILFQGGQPVKRIVGAKGKAALLRDL
MAB_4941|M.abscessus_ATCC_1997 TIAKLDVDANPETARAFQVTSIPTLILFQNGEATKRIVGAKSKSALLREL
.:**:*** ** *: ***.****:***:.* ..*******.*:****::
MSMEG_6934|M.smegmatis_MC2_155 SDAL----
TH_2412|M.thermoresistible__bu EDAL----
Mflv_0838|M.gilvum_PYR-GCK ADVV----
Mvan_6068|M.vanbaalenii_PYR-1 SEVV----
MMAR_5478|M.marinum_M SDAVPNLG
MUL_5067|M.ulcerans_Agy99 SDAVPNLG
MAV_5302|M.avium_104 SDAVPNLG
Mb3945|M.bovis_AF2122/97 SDVVPNLN
Rv3914|M.tuberculosis_H37Rv SDVVPNLN
MLBr_02703|M.leprae_Br4923 SDVVPNLN
MAB_4941|M.abscessus_ATCC_1997 DGVV----
.: