For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GQVNTLTDPSAPEGPTGQLVNWVRTLEWDQVPEHVRVRAAHLLLDGIGCALVGAQLPWSRLATDAVLGIE GDGAVPVIGTGRTSTPVGAVLLNSTFIQGFELDDFHPFAPLHSASLVVPALLATTAHLNRPVSGKELLMA AIVGFETGPRIGRALGGTEMLSRGWHSGPVFGGIGTALACGRLRGLNGEQLEDAVGFAATQSAGLMSAQY EAMGKRMQHGFAARNGFYSAALAQSGYTGIDQVLERPYGGFLAVYGEGHRPDASAITRGLGDEWETTAIM VKSWAVMGGLHGVVEAARILRNRLHGRTIEHIDIRVGDVVYHHGWWQPQRPLTAIGAQMNIGYAAAVTLL DGVALPQQFTAERLDADDVWRLLARTHVELDESIDELPPTERFQTHLTLTFSDGSTETASVMAPHGNPRD PVTNHEVVDKFARLVAPVMPADRAAAIQHAFLGLPEVPDVAPLVELLSGPVGRVLD*
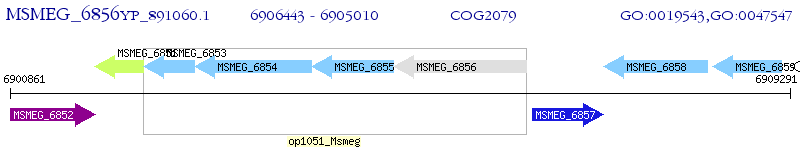
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6856 | - | - | 100% (477) | MmgE/PrpD family protein |
| M. smegmatis MC2 155 | MSMEG_4124 | - | 7e-09 | 27.18% (195) | hypothetical protein MSMEG_4124 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1742 | - | 1e-166 | 61.68% (475) | MmgE/PrpD family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6856|M.smegmatis_MC2_155 --MGQVNTLTDPSAPEGPTGQLVNWVRTLEWDQVPEHVRVRAAHLLLDGI
MAV_1742|M.avium_104 MTALQQETVTDP---VGPTGLLATWVAELTLDDVPPPVVDRAKHLLLDGI
* :*:*** **** *..** * *:** * ** *******
MSMEG_6856|M.smegmatis_MC2_155 GCALVGAQLPWSRLATDAVLGIEGDGAVPVIGTGRTSTPVGAVLLNSTFI
MAV_1742|M.avium_104 GCALVGAQLPWSRIATDAVLALEGSGDSIVIGTGRRTSAPAAAVLNGTFI
*************:******.:**.* ****** ::. .*.:**.***
MSMEG_6856|M.smegmatis_MC2_155 QGFELDDFHPFAPLHSASLVVPALLATTAHLNRPVSGKELLMAAIVGFET
MAV_1742|M.avium_104 QGFELDDFHPLAPLHSCSLLIPALLSTAATRSATTTGRELLPAAIAGFEV
**********:*****.**::****:*:* . ..:*:*** ***.***.
MSMEG_6856|M.smegmatis_MC2_155 GPRIGRALGGTEMLSRGWHSGPVFGGIGTALACGRLRGLNGEQLEDAVGF
MAV_1742|M.avium_104 GPRVGHALHGTQMLDRGWHSGPVFGTHAAAMASGKLRGLPPAQLEDALGL
***:*:** **:**.********** .:*:*.*:**** *****:*:
MSMEG_6856|M.smegmatis_MC2_155 AATQSAGLMSAQYEAMGKRMQHGFAARNGFYSAALAQSGYTGIDQVLERP
MAV_1742|M.avium_104 AGTQSAGLMAAQYEAMSKRMHHGLAARNGFYAAGLAAAGYTGIKRVFERE
*.*******:******.***:**:*******:*.** :*****.:*:**
MSMEG_6856|M.smegmatis_MC2_155 YGGFLAVYGEGHRPDASAITRGLGDEWETTAIMVKSWAVMGGLHGVVEAA
MAV_1742|M.avium_104 YGGFLSVFGEGHDPDAAALTADLGQRWETSLIMVKSYAAMGGLHGAIDAA
*****:*:**** ***:*:* .**:.***: *****:*.******.::**
MSMEG_6856|M.smegmatis_MC2_155 RILRNRLHGRTIEHIDIRVGDVVYHHGWWQPQRPLTAIGAQMNIGYAAAV
MAV_1742|M.avium_104 RRLRNSVAPQNISSVDITVGETVYKHGWWLPERPLTPIGAQMNIGYATAA
* *** : :.*. :** **:.**:**** *:****.**********:*.
MSMEG_6856|M.smegmatis_MC2_155 TLLDGVALPQQFTAERLDADDVWRLLARTHVELDESIDELPPTERFQTHL
MAV_1742|M.avium_104 ALLDGNVLPEQFTAARLDADDIWALISATRVHLDESLADADITEKFRTDL
:**** .**:**** ******:* *:: *:*.****: : **:*:*.*
MSMEG_6856|M.smegmatis_MC2_155 TLTFSDGSTETASVMAPHGNPRDPVTNHEVVDKFARLVAPVMPADRAAAI
MAV_1742|M.avium_104 AVTTREGTVHRARVALPHGAPNDPVTNDEVVAKFHALADRVTSRGRAAAI
::* :*:.. * * *** *.*****.*** ** *. * . .*****
MSMEG_6856|M.smegmatis_MC2_155 QHAFLGLPEVPDVAPLVELLSGPVGRVLD
MAV_1742|M.avium_104 ERAVIRLDDLTDVENLMDLLADPVAGALD
::*.: * ::.** *::**:.**. .**