For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALKIVIGGDNAGFNYKEALRKDLEADDRVASVEDVGVGGVDDTTSYPNVAVAAAEKVARGEADRALLICG TGLGVAIAANKVKGIRAVTAHDVYSVQRSVLSNNAQVLCMGERVVGLELARALVKEWLGLEFDPQSASAA KVNDICAYEGA*
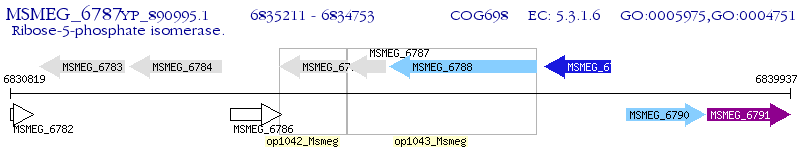
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6787 | - | - | 100% (152) | ribose 5-phosphate isomerase |
| M. smegmatis MC2 155 | MSMEG_3272 | - | 1e-57 | 71.71% (152) | ribose 5-phosphate isomerase |
| M. smegmatis MC2 155 | MSMEG_4684 | - | 4e-14 | 30.41% (148) | ribose-5-phosphate isomerase B |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2492c | - | 2e-10 | 25.68% (148) | ribose-5-phosphate isomerase B |
| M. gilvum PYR-GCK | Mflv_2581 | - | 9e-13 | 27.70% (148) | ribose-5-phosphate isomerase B |
| M. tuberculosis H37Rv | Rv2465c | - | 2e-10 | 25.68% (148) | ribose-5-phosphate isomerase B |
| M. leprae Br4923 | MLBr_01484 | - | 2e-09 | 23.65% (148) | ribose-5-phosphate isomerase B |
| M. abscessus ATCC 19977 | MAB_1574 | - | 2e-11 | 27.15% (151) | ribose-5-phosphate isomerase B |
| M. marinum M | MMAR_3813 | rpiB | 7e-11 | 26.35% (148) | ribose/galactose isomerase RpiB |
| M. avium 104 | MAV_1707 | - | 1e-11 | 27.70% (148) | ribose-5-phosphate isomerase B |
| M. thermoresistible (build 8) | TH_0347 | rpi | 4e-13 | 28.38% (148) | PROBABLE ISOMERASE |
| M. ulcerans Agy99 | MUL_3738 | rpiB | 1e-10 | 25.68% (148) | ribose-5-phosphate isomerase B |
| M. vanbaalenii PYR-1 | Mvan_4053 | - | 2e-12 | 27.70% (148) | ribose-5-phosphate isomerase B |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01484|M.leprae_Br4923 MSGMRVYLGADHAGYELKRQIIEHLKQSGHQP--IDCGAFSYDVDDEYPA
MAV_1707|M.avium_104 ---MRVYLGSDHAGFELKQQIIAHLEQSGHQP--IDCGAFSYDADDDYPA
Mb2492c|M.bovis_AF2122/97 MSGMRVYLGADHAGYELKQRIIEHLKQTGHEP--IDCGALRYDADDDYPA
Rv2465c|M.tuberculosis_H37Rv MSGMRVYLGADHAGYELKQRIIEHLKQTGHEP--IDCGALRYDADDDYPA
MMAR_3813|M.marinum_M ---MRVYLGADHAGLELKQQIIEHLKATGHEP--IDCGAFTYDAEDDYPA
MUL_3738|M.ulcerans_Agy99 ---MRVYLGADHAGLELKQQIIEHLKATGHEP--IDCGAFTYDAEDDYPA
TH_0347|M.thermoresistible__bu ---MRVYLGADHAGFELKQKIIEHLRENGHEP--VDCGAYTYDADDDYPP
Mflv_2581|M.gilvum_PYR-GCK ---MRVYVGADHAGYELKQAIVEHLAATGHEP--VDCGAFAYDAEDDYPA
Mvan_4053|M.vanbaalenii_PYR-1 ---MRVYVGADHAGYELKQTIVEHLKATGHEP--VDCGAFTYDAEDDYPA
MAB_1574|M.abscessus_ATCC_1997 ---MRVYVGADHAGYELKQKIIEHLRGRGHEP--VDCGAFELDPQDDYPA
MSMEG_6787|M.smegmatis_MC2_155 -MALKIVIGGDNAGFNYKEALRKDLEADDRVASVEDVGVGGVDDTTSYPN
::: :*.*:** : *. : .* .: . * *. * .**
MLBr_01484|M.leprae_Br4923 FCITAATRTVADPGSLGIVLGGSGNGEQIAANKVVGARCALAWSVETARL
MAV_1707|M.avium_104 FCIAAATRTVADPDSLGIVLGGSGNGEQIAANKVPGARCALAWSVETAQL
Mb2492c|M.bovis_AF2122/97 FCIAAATRTVADPGSLGIVLGGSGNGEQIAANKVPGARCALAWSVQTAAL
Rv2465c|M.tuberculosis_H37Rv FCIAAATRTVADPGSLGIVLGGSGNGEQIAANKVPGARCALAWSVQTAAL
MMAR_3813|M.marinum_M FCIAAAQRTVADPGSLGIVLGGSGNGEQIAANKVPGARCALAWSEQTASL
MUL_3738|M.ulcerans_Agy99 FCIAAAQRTVADPGSLGIVLGGSGNGEQIAANKVPGARCALVWSEQTASL
TH_0347|M.thermoresistible__bu FCIAAAERTVADPGSLGIVLGGSGNGEQIAANKVPGARCALAWSVETAKL
Mflv_2581|M.gilvum_PYR-GCK FCIAAAEKTVADPGSLGIVLGGSGNGEQIAANKVPGARCALAWSTETATL
Mvan_4053|M.vanbaalenii_PYR-1 FCIAAAQKTVADPGSLGIVLGGSGNGEQIAANKVPGARCALAWSPETASL
MAB_1574|M.abscessus_ATCC_1997 FCIDAARRTVADPGSLGIVLGGSGNGEQIAANKVPGARCALAWSLETAVL
MSMEG_6787|M.smegmatis_MC2_155 VAVAAAEKVARGEADRALLICGTGLGVAIAANKVKGIRAVTAHDVYSVQR
..: ** :.. . . .::: *:* * ****** * *.. . . :.
MLBr_01484|M.leprae_Br4923 AREHNNAQLIGIGGRMHTVAEALTIVDAFVTTPWSEAQRHQRRIDILAEF
MAV_1707|M.avium_104 AREHNNAQLIGIGGRMHTVAEALAIVDAFVTTPWSKAPRHQRRIDILAEY
Mb2492c|M.bovis_AF2122/97 AREHNNAQLIGIGGRMHTVAEALAIVDAFVTTPWSKAQRHQRRIDILAEY
Rv2465c|M.tuberculosis_H37Rv AREHNNAQLIGIGGRMHTVAEALAIVDAFVTTPWSKAQRHQRRIDILAEY
MMAR_3813|M.marinum_M AREHNNAQLIGIGGRMHTVAEALAIVDAFLSTPWSKAERHQRRIDILSEY
MUL_3738|M.ulcerans_Agy99 AREHNNAQLIGIGGRMHTVAEALAIVDAFLSTPWSKAERHQRRIDILSEY
TH_0347|M.thermoresistible__bu AREHNNAQLIGIGGRMHTVEEALAIVDAFLSTPWSEAERHQRRIDILSEY
Mflv_2581|M.gilvum_PYR-GCK AREHNNAQLVGIGGRMHSTDEALAIVDAFLAAKWSEAPRHQRRIDILAEF
Mvan_4053|M.vanbaalenii_PYR-1 AREHNNAQLIGIGGRMHSTEEALAIVDAFLTTKWSEAPRHQRRIDILAEY
MAB_1574|M.abscessus_ATCC_1997 AREHNNAQLIGIGGRMHTVPEALAIVDAFLAAKWSEEPRHQRRIDILAEY
MSMEG_6787|M.smegmatis_MC2_155 SVLSNNAQVLCMGERVVGLELARALVKEWLGLEFDPQSASAAKVNDICAY
: ****:: :* *: * ::*. :: :. ::: :. :
MLBr_01484|M.leprae_Br4923 ERTHQAPPVPGAQA
MAV_1707|M.avium_104 ERTHQAPPVPGAVG
Mb2492c|M.bovis_AF2122/97 ERTHEAPPVPGAPA
Rv2465c|M.tuberculosis_H37Rv ERTHEAPPVPGAPA
MMAR_3813|M.marinum_M ERTHQAPPVPGAQD
MUL_3738|M.ulcerans_Agy99 ERTHQAPPVPGAQD
TH_0347|M.thermoresistible__bu ERTRQAPPVPGAPA
Mflv_2581|M.gilvum_PYR-GCK EKTHVAPAVPGA--
Mvan_4053|M.vanbaalenii_PYR-1 EKTHVAPPVPGA--
MAB_1574|M.abscessus_ATCC_1997 ETTRVAPPVPGASS
MSMEG_6787|M.smegmatis_MC2_155 EGA-----------
* :