For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MEHPGETTLQGRSGMSGRPWGSAHVGQTIPTVAAEFMSAQRLAVVGARDDAGRVWAGPLTGPPGFLSARG ETTVVAAAVPGPLDPLAGAFDTGRAVGMLIIDFTSRKRMRVNGIATRDGESLIVETDQVYANCPKYIQMR EPADAPALPPPSRTVGAELTAAQRVWIESSDTFFIATQADPYGTDASHRGGHPGFVTTSSACRLSWPEYS GNLMYMTLGNLEVDSRAGLLFVDWERGSTLHLTGHARTDWERQVVDFDLEQVVEIDAAIPLTWTFVEYSK FNPA
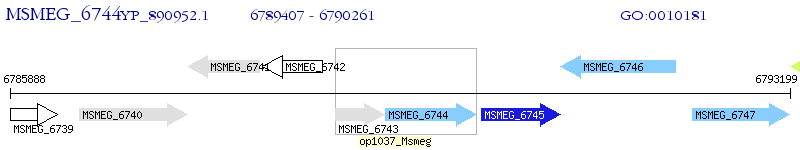
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6744 | - | - | 100% (284) | oxidoreductase FAD/NAD |
| M. smegmatis MC2 155 | MSMEG_1061 | - | 1e-05 | 31.52% (92) | phosphohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0034 | - | 2e-05 | 30.09% (113) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3055c | - | 4e-66 | 46.94% (294) | hypothetical protein MAB_3055c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4731 | - | 2e-09 | 30.34% (89) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6744|M.smegmatis_MC2_155 -MEHPGETTLQ--GRSGMSGR----------PWGS-AHVGQTIPTVAAEF
MAB_3055c|M.abscessus_ATCC_199 MAYHPGELAVQ--HRMGQSDN----------AIRVGQIISADIPEVAAAF
Mflv_0034|M.gilvum_PYR-GCK -MSRHVTTVEE--LRAIVGHP----------HAVVAGKVRDRLLPAMRDW
Mvan_4731|M.vanbaalenii_PYR-1 MSKHYGSIAFTGDVRAVQSEHGSEAFYGRKLAAGKASSGRDPLTGEEKEY
: . * . : :
MSMEG_6744|M.smegmatis_MC2_155 MSAQRLAVVGARDDAGRVWAGPLTGPPGFLSARGETTVVAAAVPG--PLD
MAB_3055c|M.abscessus_ATCC_199 LAAQPLVFISARDDDGRMWASQIVGPPGFAHAADARTIVIDATPD--YGD
Mflv_0034|M.gilvum_PYR-GCK LAHSPLGFVATTDADGRVDVSPKGDPAGFVHIIDDHTIAIPERPGNKRVD
Mvan_4731|M.vanbaalenii_PYR-1 LSERDGLFLASVGETGWPYVQFRGGPRGFVRVLDDHTIGWADFRGNLQYI
:: .:.: . * . .* ** . *: .
MSMEG_6744|M.smegmatis_MC2_155 PLAGAFDTGRAVGMLIIDFTSRKRMRVNGIATRDGESLIVETDQVYANCP
MAB_3055c|M.abscessus_ATCC_199 PLAGVLQSERKIGMLALQPQRRRRMRVNGTSVPSGDGLIVTTDQVYSNCP
Mflv_0034|M.gilvum_PYR-GCK GYLNVLERPR-VGTLFVIPGRGDTLRINGSAR-------VMADADYFDAM
Mvan_4731|M.vanbaalenii_PYR-1 STGNVNGDDR-VAIIAVDYAQRRRLKIFGHAR----IVTAEQDAALIQSL
.. * :. : : ::: * : . * :.
MSMEG_6744|M.smegmatis_MC2_155 KYIQMREPADA-PALPPPSRTVGAELTAAQRVWIESSDTFFIATQADPYG
MAB_3055c|M.abscessus_ATCC_199 KYIARRDISEIGPSLPGETR-RGSALDDAQQQAVSVTDTFIIGTADGDGN
Mflv_0034|M.gilvum_PYR-GCK SVEGRRPILAL--------------EIAVEEVFFHCAKAFLRSDAWDPQS
Mvan_4731|M.vanbaalenii_PYR-1 SDSTYDAVVER--------------AVVVGVDAYDWNCPQHITPRFTLEE
. . .
MSMEG_6744|M.smegmatis_MC2_155 TDASHRGGHPGFVTTSSACRLSWPEYSGNLMYMTLGNLEVDSRAGLLFVD
MAB_3055c|M.abscessus_ATCC_199 ADASHRGGNPGFLRLLSPKLLRWPDYLGNSMFMTLGNLHVDPRCGLLIPD
Mflv_0034|M.gilvum_PYR-GCK WDP---------AAVPSAAHLAKQMYADADLDDMRRALDEDMMRQALY--
Mvan_4731|M.vanbaalenii_PYR-1 LEP---------ALAPLRSELSSLQAENARLREQLERAGA----------
:. * . :
MSMEG_6744|M.smegmatis_MC2_155 WERGSTLHLTGHARTDWER--------QVVDFDLEQVVEIDAAIPLTWTF
MAB_3055c|M.abscessus_ATCC_199 WRTGSTLQLTGTARLNWDESTFAPGAQCSVDFTIDEVVETVNGSPLRWGH
Mflv_0034|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4731|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6744|M.smegmatis_MC2_155 VEYSKFNPA-
MAB_3055c|M.abscessus_ATCC_199 AEPSPANPPL
Mflv_0034|M.gilvum_PYR-GCK ----------
Mvan_4731|M.vanbaalenii_PYR-1 ----------