For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAGFALGHAGINVTDLSRSKSFYQTVFGFDVLREHTDGDKRFAFLGTDGTLAVTLWEQSSGVFATDRPGL HHLAFLVDDIDAVRAAEARVRDAGAEVLHGGVVPHQEGASSGGIFFTDPDGTRIEIYSATGADVVGAHAA PFGDAPTCGFF
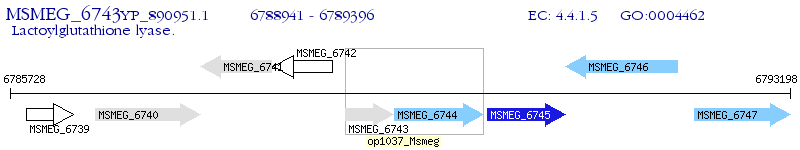
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6743 | - | - | 100% (151) | lactoylglutathione lyase |
| M. smegmatis MC2 155 | MSMEG_0884 | - | 1e-08 | 31.40% (121) | glyoxalase family protein |
| M. smegmatis MC2 155 | MSMEG_4313 | - | 1e-07 | 28.91% (128) | glyoxalase/bleomycin resistance protein/dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0135 | - | 2e-08 | 29.86% (144) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3056c | - | 2e-44 | 56.55% (145) | hypothetical protein MAB_3056c |
| M. marinum M | MMAR_4129 | gloA_2 | 2e-08 | 31.82% (132) | glyoxalase, GloA_2 |
| M. avium 104 | MAV_1440 | - | 1e-09 | 33.09% (136) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. thermoresistible (build 8) | TH_3357 | - | 4e-08 | 31.76% (148) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_3992 | gloA_2 | 1e-08 | 31.82% (132) | glyoxalase, GloA_2 |
| M. vanbaalenii PYR-1 | Mvan_0768 | - | 5e-08 | 31.67% (120) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6743|M.smegmatis_MC2_155 MAG-----FALGHAGINVTDLSRSKSFYQTVFGFDVL--REHTDGD----
MAB_3056c|M.abscessus_ATCC_199 MSAATAPRVVTGHIGLNVSDLERSIAFYRQAFGFDEL--AVSADGA----
MMAR_4129|M.marinum_M ---MGVSSTSIGHVRLTVTDIERSRRFYESVFEWPVL--IELPANADEAT
MUL_3992|M.ulcerans_Agy99 MGRIGVSSTSIGHVRLTVTDIERSRRFYESVFEWPVL--IELPANADEAT
MAV_1440|M.avium_104 ---MGLTNTSIAHVRLTVTDIERSRRFYESVFGWPVL--LEVPDNADAST
Mflv_0135|M.gilvum_PYR-GCK ----MYGGTVIDHFGINCADWERSKAFYDKVLGVLGY--TRQMDFG----
Mvan_0768|M.vanbaalenii_PYR-1 ---------MIDHFGINCADWETSKAFYDKVLGVLGY--TRQMDFQ----
TH_3357|M.thermoresistible__bu ----MSDRIRLTHFGLCVRDIGRAEQFYCNALGFEAIGRMTVDDDATATL
* : * : ** .:
MSMEG_6743|M.smegmatis_MC2_155 -KRFAFLG-----TDGTLAVTLWEQS-------SGVFATDRPGLHHLAFL
MAB_3056c|M.abscessus_ATCC_199 -QRFAFLG-----FDSGPVLTLWEQS-------SGEFSAATPGLHHLSFQ
MMAR_4129|M.marinum_M RRQLAFLYGGVIYDLGGALLGLRPVA-------NDSFNENRVGLDHIAFR
MUL_3992|M.ulcerans_Agy99 RRQLAFLYGGVIYDLGGALLGLRPVA-------NDSFNENRVGLDHIAFR
MAV_1440|M.avium_104 REQLSFLFGGVIYDLGGTLLGLRPVA-------TDRFDEDRTGLDHIAFR
Mflv_0135|M.gilvum_PYR-GCK ---VAIGYG----TDGHPDFWIADMS-------TGAAAGPN-REVHIAFA
Mvan_0768|M.vanbaalenii_PYR-1 ---VAVGYG----VEGKPDFWIADMS-------SGDAAGPN-REVHIAFQ
TH_3357|M.thermoresistible__bu LDIPGLLLELVYLQRDGFRLELLGYARPATVGEQGPRPMNAVGFTHLSFR
.. . . : : . *::*
MSMEG_6743|M.smegmatis_MC2_155 VDDIDAVRAAEARVRDAGAEVLHGGVVPHQEGASSGGIFFTDPDGTRIEI
MAB_3056c|M.abscessus_ATCC_199 VDSVRQVQQVEAILKQLSTVFVHDGVVAHREGATSGGIFFTDPDGIRLEV
MMAR_4129|M.marinum_M VGSIDELNSAAAHLDDLG--IAHEPVKDIGP---SYILEFRDPDNIALEL
MUL_3992|M.ulcerans_Agy99 VGSIDELNSAAAHLDDLG--IAHEPVKDIGP---SYILEFRDPDNIALEL
MAV_1440|M.avium_104 LDSRDELDAAAAHLDELG--VAHEPVKDIGP---AYILQFRDPDNIALEL
Mflv_0135|M.gilvum_PYR-GCK AKDAESVQAFFRTALALGVEPLHEPRLWPEYHENYYGAFVRDPDGNNVEA
Mvan_0768|M.vanbaalenii_PYR-1 AAGVEAVQAFYDAALDAGAESLHAPRLWPEYHPGYYGAFVRDPDGNNVEA
TH_3357|M.thermoresistible__bu VDDAD--RVMDAIVAHGGRTLPERTVAFSGG---NRGLMATDPEGNLLEL
. . . **:. :*
MSMEG_6743|M.smegmatis_MC2_155 YSATGADVVGAHAAPFGDAPTCGFF
MAB_3056c|M.abscessus_ATCC_199 FAASGAEQ---HQAPVGDAPTCGFF
MMAR_4129|M.marinum_M TAPK---------------------
MUL_3992|M.ulcerans_Agy99 TAPK---------------------
MAV_1440|M.avium_104 TAPK---------------------
Mflv_0135|M.gilvum_PYR-GCK VFHGGGPSGGPPGPRPA--------
Mvan_0768|M.vanbaalenii_PYR-1 VFHGA--------------------
TH_3357|M.thermoresistible__bu IERTADR------------------