For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AEYTLPELDYDYAALEPHISGQINEIHHSKHHATYVKGVNDAIAKLEEARANGDHGAIFLHEKNLAFHLG GHVNHTIWWKNLSPHGGDKPTGDLAAAIDDQFGSFDNFRAQFTAAANGLQGSGWAVLGYDTLGDRLLTFQ LYDQQANVPLGIIPLLLVDMWEHAYYLQYKNVKADYVKAIWNVVNWQDVQVRFAAATGKGPDPIV*
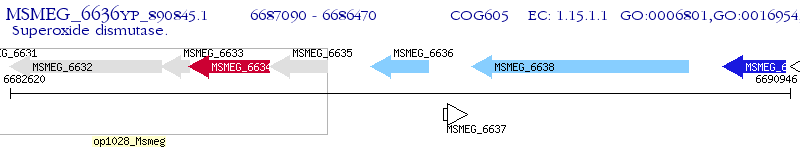
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6636 | - | - | 100% (206) | [Mn] superoxide dismutase [Mycobacterium smegmatis str. MC2 155] |
| M. smegmatis MC2 155 | MSMEG_6427 | - | e-108 | 90.00% (200) | [Mn] superoxide dismutase [Mycobacterium smegmatis str. MC2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3876 | sodA | 5e-95 | 78.00% (200) | superoxide dismutase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv3846 | sodA | 5e-95 | 78.00% (200) | superoxide dismutase |
| M. leprae Br4923 | MLBr_00072 | sodA | 1e-101 | 84.00% (200) | superoxide dismutase |
| M. abscessus ATCC 19977 | MAB_0118c | - | 1e-106 | 88.00% (200) | superoxide dismutase (Mn) |
| M. marinum M | MMAR_5396 | sodA | 4e-94 | 77.00% (200) | superoxide dismutase |
| M. avium 104 | MAV_0182 | - | 1e-106 | 89.00% (200) | [Mn]-superoxide dismutase [Mycobacterium avium 104] |
| M. thermoresistible (build 8) | TH_0750 | sodA | 1e-108 | 89.00% (200) | SUPEROXIDE DISMUTASE |
| M. ulcerans Agy99 | MUL_5018 | sodA | 3e-94 | 77.00% (200) | superoxide dismutase |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6636|M.smegmatis_MC2_155 MAEYTLPELDYDYAALEPHISGQINEIHHSKHHATYVKGVNDAIAKLEEA
TH_0750|M.thermoresistible__bu VAEYTLPDLDWDYGALEPHISGQINELHHSKHHATYVKGANDALAKLEEA
MAB_0118c|M.abscessus_ATCC_199 MAEYTLPDLDYDYGALEPHISGQINELHHSKHHATYVKGVNDAVAKLEEA
MLBr_00072|M.leprae_Br4923 MAEYTLPDLDWDYAALEPHISGEINEIHHTKHHAAYVKGVNDALAKLDEA
MAV_0182|M.avium_104 MAEYTLPDLDWDYAALEPHISGQINEIHHTKHHATYVKGVNDALAKLEEA
Mb3876|M.bovis_AF2122/97 MAEYTLPDLDWDYGALEPHISGQINELHHSKHHATYVKGANDAVAKLEEA
Rv3846|M.tuberculosis_H37Rv MAEYTLPDLDWDYGALEPHISGQINELHHSKHHATYVKGANDAVAKLEEA
MMAR_5396|M.marinum_M MAEYTLPDLDWDYGALEPHISGQINELHHSKHHATYVKGANDAVTKLEEA
MUL_5018|M.ulcerans_Agy99 MAEYTLPDLDWDYGALEPHISGQINELHHSKHHATYVKGANDAVTKLEEA
:******:**:**.********:***:**:****:****.***::**:**
MSMEG_6636|M.smegmatis_MC2_155 RANGDHGAIFLHEKNLAFHLGGHVNHTIWWKNLSPHGGDKPTGDLAAAID
TH_0750|M.thermoresistible__bu RANGDHSAIFLHEKNLAFHLGGHVNHTIWWKNLSPNGGDKPTGELAAAID
MAB_0118c|M.abscessus_ATCC_199 REKGDHAAIFLNEKNLAFHLGGHVNHSIWWKNLSPNGGDKPTGDLAAAID
MLBr_00072|M.leprae_Br4923 RAKDDHSAIFLNEKNLAFHLGGHVNHSIWWKNLSPNGGDKPTGGLATDID
MAV_0182|M.avium_104 RANEDHAAIFLNEKNLAFHLGGHVNHSIWWKNLSPDGGDKPTGELAAAID
Mb3876|M.bovis_AF2122/97 RAKEDHSAILLNEKNLAFNLAGHVNHTIWWKNLSPNGGDKPTGELAAAIA
Rv3846|M.tuberculosis_H37Rv RAKEDHSAILLNEKNLAFNLAGHVNHTIWWKNLSPNGGDKPTGELAAAIA
MMAR_5396|M.marinum_M RAKEDHSTILLNEKNLAFNLAGHVNHTIWWKNLSPNGGDKPTGELAAAID
MUL_5018|M.ulcerans_Agy99 RAKEDHSTILLNEKNLAFNLAGHVNHTIWWKNLSPNGGDKPTGELAAAID
* : **.:*:*:******:*.*****:********.******* **: *
MSMEG_6636|M.smegmatis_MC2_155 DQFGSFDNFRAQFTAAANGLQGSGWAVLGYDTLGDRLLTFQLYDQQANVP
TH_0750|M.thermoresistible__bu EHFGSFDKFRAQFTAAANGLQGSGWAVLGYDTLGQRLLTFQLYDQQANVP
MAB_0118c|M.abscessus_ATCC_199 DQFGSFDKFQAQFTAAANGLQGSGWAVLGYDSLGQKLLTFQLYDQQANVP
MLBr_00072|M.leprae_Br4923 ETFGSFDKFRAQFSAAANGLQGSGWAVLGYDTLGNKLLTFQLYDQQANVS
MAV_0182|M.avium_104 DAFGSFDKFRAQFSAAANGLQGSGWAVLGYDTLGSRLLTFQLYDQQANVP
Mb3876|M.bovis_AF2122/97 DAFGSFDKFRAQFHAAATTVQGSGWAALGWDTLGNKLLIFQVYDHQTNFP
Rv3846|M.tuberculosis_H37Rv DAFGSFDKFRAQFHAAATTVQGSGWAALGWDTLGNKLLIFQVYDHQTNFP
MMAR_5396|M.marinum_M EAFGSFDKFRAQFHAAATTVQGSGWAALGWDTLGNKLLIFQVYDHQTNFP
MUL_5018|M.ulcerans_Agy99 EAFGSFDKFRAQFHAAATTVQGSGWAALGWDTLGNKLLIFQVYDHQTNFP
: *****:*:*** ***. :******.**:*:**.:** **:**:*:*..
MSMEG_6636|M.smegmatis_MC2_155 LGIIPLLLVDMWEHAYYLQYKNVKADYVKAIWNVVNWQDVQVRFAAATGK
TH_0750|M.thermoresistible__bu LGIIPLLQVDMWEHAFYLQYKNVKADYVKAFWNVVNWEDVQNRFNAAVSK
MAB_0118c|M.abscessus_ATCC_199 LGIIPLLQVDMWEHAFYLQYKNVKADYVKAFWNVVNWADVQDRYTAATTK
MLBr_00072|M.leprae_Br4923 LGIIPLLQVDMWEHAFYLQYKNVKADYVKAFWNVVNWADVQSRYMAATSK
MAV_0182|M.avium_104 LGIIPLLQVDMWEHAFYLQYKNVKADYVKAFWNVVNWADVQKRYAAATSK
Mb3876|M.bovis_AF2122/97 LGIVPLLLLDMWEHAFYLQYKNVKVDFAKAFWNVVNWADVQSRYAAATSQ
Rv3846|M.tuberculosis_H37Rv LGIVPLLLLDMWEHAFYLQYKNVKVDFAKAFWNVVNWADVQSRYAAATSQ
MMAR_5396|M.marinum_M LGIVPLLLLDMWEHAFYLQYKNVKVDFAKAFWNVVNWADVQSRYDAATSK
MUL_5018|M.ulcerans_Agy99 LGIVPLLLLDMWEHAFYLQYKNVKVDFAKAFWNVVNWADVQSRYDAATSK
***:*** :******:********.*:.**:****** *** *: **. :
MSMEG_6636|M.smegmatis_MC2_155 GPDPIV-
TH_0750|M.thermoresistible__bu AQGLIF-
MAB_0118c|M.abscessus_ATCC_199 TSGLIFG
MLBr_00072|M.leprae_Br4923 TQGLIFD
MAV_0182|M.avium_104 AQGLIFG
Mb3876|M.bovis_AF2122/97 TKGLTFG
Rv3846|M.tuberculosis_H37Rv TKGLIFG
MMAR_5396|M.marinum_M TQGLIFG
MUL_5018|M.ulcerans_Agy99 TQGLIFG
. .