For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSDRTALLAHGLGGSTDLPIPYTYALLGAAWTLTFTFAVVAFAWRRPRFDPDKPGHPLPEWVTTFVDSP VTRWGAGLLALAFTGWVGALSFTGPQDASNPLPGVFYVLLWVGLVAVSLLIGPVWRLISPVRTLIRLIPA TTGRLRYPARLGYWPAAAGLFAFVWLELASADPGSLAAIRNWLLIYLAVTLAGALCCGQRWLSNADPFEV YSTAASKLSPLRRHAGRIVIGNPFDHLRSLPVRPGVVAVLAVLLGSTAFDSFSAMPVWRNFVDDNSGSAS GATLIRTAGLAVFVATVALTFWAAARATGGVDPQQRRELPGQMAHSLVPIVIGYVFAHYLTYLVERGQQT IFLLFGMHDAQINYILSMHPAVLSSLKVGFVLAGHVAGVIAAHDRALRLLPARHRLTGQLAMMLVMVGYT FTGLYLLFGG
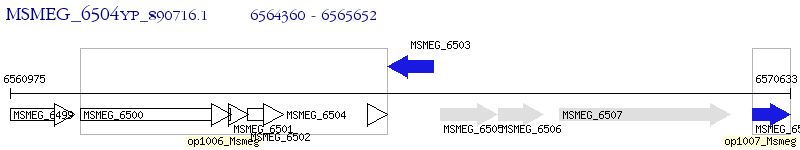
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6504 | - | - | 100% (430) | hypothetical protein MSMEG_6504 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1091 | - | 1e-174 | 70.75% (424) | hypothetical protein Mflv_1091 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1629c | - | 1e-170 | 67.57% (441) | hypothetical protein MAB_1629c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3200 | - | 1e-157 | 63.23% (446) | hypothetical protein MAV_3200 |
| M. thermoresistible (build 8) | TH_1794 | - | 0.0 | 72.02% (436) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5726 | - | 0.0 | 73.76% (423) | hypothetical protein Mvan_5726 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1091|M.gilvum_PYR-GCK ----MTAQILAHGLGGSTDLPIPFTYALVGAAWTLTATFAVVAFAWRTPK
Mvan_5726|M.vanbaalenii_PYR-1 ---------MAHGLGGSADLPIPYTYALIGAAWALTFTFAVVALAWRTPR
MAB_1629c|M.abscessus_ATCC_199 MTGDRTTTVLAHGLGGSADLPIPYTYALIGAAWALTFTFAVVALAWRKPR
TH_1794|M.thermoresistible__bu ------VTYLAHGIGGSTDLPIPFTYALIGASWALAITFAVVAFAWRRPR
MSMEG_6504|M.smegmatis_MC2_155 -MTSDRTALLAHGLGGSTDLPIPYTYALLGAAWTLTFTFAVVAFAWRRPR
MAV_3200|M.avium_104 MSPAPAAVVLAHGLGGSGDLPVPYAYAMVGAAWALTFTFALVAFAWRRPR
:***:*** ***:*::**::**:*:*: ***:**:*** *:
Mflv_1091|M.gilvum_PYR-GCK FDAARPGRALPPWVTRSVDAPATRWVLAAAALGLTGWVAVAAVRGPQTSE
Mvan_5726|M.vanbaalenii_PYR-1 FDAARPGRPLPQWVTRAVDAPATRWTLAAATLLFTGWVAVSTFVGPQDSG
MAB_1629c|M.abscessus_ATCC_199 FDTNRTGRPLPCWVTAIVDAPAARGAVTAVVLAATAWITVAAVLGPQDGR
TH_1794|M.thermoresistible__bu FDPDRPGRALPRWVTTVVDAAATRWALAAAALLFTGWVALAAFFGPQDRS
MSMEG_6504|M.smegmatis_MC2_155 FDPDKPGHPLPEWVTTFVDSPVTRWGAGLLALAFTGWVGALSFTGPQDAS
MAV_3200|M.avium_104 FDPLKPGRALPAALTAFVDARATRYTAAGLALAVAAWAVAAGVWGPESDA
**. :.*:.** :* **: .:* .* :.* . **:
Mflv_1091|M.gilvum_PYR-GCK NPLPGVFYVLLWVGVVALSLAIGPVWRVLSPVRALH----RFL--GGRNL
Mvan_5726|M.vanbaalenii_PYR-1 NALPGVFYVLLWVGLVALSVLLGPVWRVLSPVRALH----RFS--GGRSL
MAB_1629c|M.abscessus_ATCC_199 NALPGVFYVLLWVGVVAASLLCGPVWRVLSPVRAAY----RLLPLARRPP
TH_1794|M.thermoresistible__bu NPLPGVFYVLLWVGLVALSALIGPVWRAISPVRAVY----RLLSLPRRLP
MSMEG_6504|M.smegmatis_MC2_155 NPLPGVFYVLLWVGLVAVSLLIGPVWRLISPVRTLI----RLIP--ATTG
MAV_3200|M.avium_104 NALLGAFYVLLWVGLVAVSLAIGPVWRVISPIRTLYLLARRVVPERLARP
*.* *.********:** * ***** :**:*: *.
Mflv_1091|M.gilvum_PYR-GCK GRRYPDALGYWPAAAGLFAFVWLELASPDPGSLGAIKIWLVVYLAVTLAG
Mvan_5726|M.vanbaalenii_PYR-1 GRTYPAALGYWPAAAGLFAFVWLELASPDPGSLGAIRTWLLVYLAVTVAG
MAB_1629c|M.abscessus_ATCC_199 PVAYPRRWGYWPAAAGLYLFVWLELASADPGSLGAVKIWLLGYLCVTMAG
TH_1794|M.thermoresistible__bu DRGYPRQLGYWPAALGLFAFVWLELAGPNPGSLTAIRTWLLVYLVVTLAG
MSMEG_6504|M.smegmatis_MC2_155 RLRYPARLGYWPAAAGLFAFVWLELASADPGSLAAIRNWLLIYLAVTLAG
MAV_3200|M.avium_104 RLSYPKRWGYRPAALGLFAFVWMELASPNPAAPSWVTGWLLVYAVLMAGG
** ** *** **: ***:***..:*.: : **: * : .*
Mflv_1091|M.gilvum_PYR-GCK ALWCGPRWNARADPFEVYSVVASRFSLFGRNPD-GRIAIGNPFNRLLTLP
Mvan_5726|M.vanbaalenii_PYR-1 ALWCGTQWCARADPFEVYSVVASRVSPLRRNSD-GRIAIGNPFDHLPSLP
MAB_1629c|M.abscessus_ATCC_199 ALACGTRWCAQADPFEVYSMVASRLSPLRRNTD-GLIAYGNPFDHLPSLP
TH_1794|M.thermoresistible__bu ALWCGPRWFGRADPFEVYSVVASRVSALRRNPQSGRIVIGNPFDHLPSLP
MSMEG_6504|M.smegmatis_MC2_155 ALCCGQRWLSNADPFEVYSTAASKLSPLRRHAG--RIVIGNPFDHLRSLP
MAV_3200|M.avium_104 AWLCGQRWLARADPFGVYSMAVSRLSPFRRNPRTGRIVVGNPLDHLPSLP
* ** :* ..**** *** ..*:.* : *:. *. ***:::* :**
Mflv_1091|M.gilvum_PYR-GCK VRPGVVAVLSVLLGSTAFDSFSAMPQIRRLVD----ELTD-SALAATALR
Mvan_5726|M.vanbaalenii_PYR-1 VRPGIVAVLSVLLGSTAFDSFSATPQFRGFVD----DLTD-SGLGATAVR
MAB_1629c|M.abscessus_ATCC_199 IRPGMVAVLAVLLGSTAFDSFSAMPEWRGLVD----ALAQGSTVASVAIR
TH_1794|M.thermoresistible__bu VRPGIVAVLAVLLGSTAFDSFSATPRWRDFVD----STTS-GVLTATLLN
MSMEG_6504|M.smegmatis_MC2_155 VRPGVVAVLAVLLGSTAFDSFSAMPVWRNFVD----DNSG-SASGATLIR
MAV_3200|M.avium_104 VRPGVVVLLAVLLGSTAFDSFSSSPTWRGFSDRLTREFAAPPALASSVLR
:***:*.:*:************: * * : * : : :.
Mflv_1091|M.gilvum_PYR-GCK TVGLVLFVGIVAATFTLAARCTGGVDARTRRELPGLLAHSLIPIVLGYVF
Mvan_5726|M.vanbaalenii_PYR-1 TAGLALFACVVAATFWAAARCTGGVDARRRRELPGLFAHSLIPIVIGYVF
MAB_1629c|M.abscessus_ATCC_199 TAGLTVFVIVVATTFCLAARATGGVDRATRRNLPGLLAHSLIPIVIGYIF
TH_1794|M.thermoresistible__bu TAGLLVFIGVVAGTFWGAARATGGVDPEQRRRLPGRMAHSLIPIVIGYVF
MSMEG_6504|M.smegmatis_MC2_155 TAGLAVFVATVALTFWAAARATGGVDPQQRRELPGQMAHSLVPIVIGYVF
MAV_3200|M.avium_104 TLGLIVFICVVALTFSLAARATGGVDAQQRRALPGQMAHSLIPIVVGYIF
* ** :* ** ** ***.***** ** *** :****:***:**:*
Mflv_1091|M.gilvum_PYR-GCK AHYLTYLIERGQQTVYRLFG----------SADAEVVYVLSMHPGLLASL
Mvan_5726|M.vanbaalenii_PYR-1 AHYLTYLVERGQQTVYRLLG----------QPDAEVMYVLSMHPAVLATL
MAB_1629c|M.abscessus_ATCC_199 AHYLTYLVERGQQTVLRLADPLGRGW----LADVDVSYALSMHPSVLATL
TH_1794|M.thermoresistible__bu AHYLSYLVERGQQTLYQLADPLGAGWNPFGLADTEVNYVLSMHPAVLSTL
MSMEG_6504|M.smegmatis_MC2_155 AHYLTYLVERGQQTIFLLFG----------MHDAQINYILSMHPAVLSSL
MAV_3200|M.avium_104 AHYLSYLVERGQQAVFALADPFGRGWNPLGLAHLQVAYVLSMHPPVLAAI
****:**:*****:: * . . :: * ***** :*:::
Mflv_1091|M.gilvum_PYR-GCK KVGFVLFGHVAGVIAAHDRALAVLPRQHQLTGQLAMMLVMVGYTFTGLYL
Mvan_5726|M.vanbaalenii_PYR-1 KVGFVVAGHIAGVVAAHDRALKVLPARHQLTGQLAMMLVMVGYTFTGLYL
MAB_1629c|M.abscessus_ATCC_199 KVGFVLAGHIAGVVAAHDQALKVLPKQHQVTGQLAMMLVMVGYTFTGLYL
TH_1794|M.thermoresistible__bu KVGFVVAGHIAAVVAAHDCALKLLPRAHQLTGQLAMMLVMVGYTFTGLYL
MSMEG_6504|M.smegmatis_MC2_155 KVGFVLAGHVAGVIAAHDRALRLLPARHRLTGQLAMMLVMVGYTFTGLYL
MAV_3200|M.avium_104 KVTCVVTGHIVAVIAAHDKALRLLPAGHQLTGQLAMMLVMVGYTFTGLYL
** *: **:..*:**** ** :** *::********************
Mflv_1091|M.gilvum_PYR-GCK LFGG
Mvan_5726|M.vanbaalenii_PYR-1 LFGG
MAB_1629c|M.abscessus_ATCC_199 LFGG
TH_1794|M.thermoresistible__bu LFGG
MSMEG_6504|M.smegmatis_MC2_155 LFGG
MAV_3200|M.avium_104 LFGG
****