For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSHADTALPPRAERLFALAEQVTGFMPVDEGRALYDAAVRYLGGGVGVEIGTYCGKSTVMLGAAAQETGS VLYTVDHHHGSEEHQPGWEYHDASLVDPVTGLFDTLPKMRHTLDAAGLDDHVVAVVGRSPVVARGWRTPL RFLFIDGGHTEEAAQRDFDGWARWVQKGGALVIHDVFPNPDDGGQAPFHIYQRALATGAFREVSATGSMR VLERISGEAGTLD
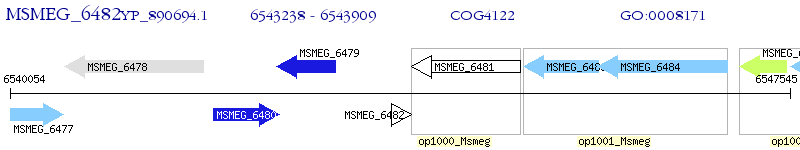
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6482 | - | - | 100% (223) | secreted protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1102 | - | 1e-106 | 81.08% (222) | hypothetical protein Mflv_1102 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2922 | - | 1e-96 | 73.24% (213) | O-methyltransferase |
| M. avium 104 | MAV_2758 | - | 1e-100 | 77.27% (220) | secreted protein |
| M. thermoresistible (build 8) | TH_0085 | - | 1e-110 | 82.88% (222) | secreted protein |
| M. ulcerans Agy99 | MUL_1691 | - | 4e-05 | 27.38% (168) | O-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5715 | - | 1e-107 | 82.65% (219) | hypothetical protein Mvan_5715 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2922|M.marinum_M MSS-----DIPSAVQRIFALADQVPGFMPADEGEALFNAALRYFNGGLGV
MAV_2758|M.avium_104 ---------MNHVDERLFALAEQVQGFMPAEEGRALYDTALRYLDGGTGV
Mflv_1102|M.gilvum_PYR-GCK MDT--TDAALPPAAAALFEFADTVIGFMPADEGRALYDTAVQYLADGVGV
Mvan_5715|M.vanbaalenii_PYR-1 MASRQSDTALPPVAAALFELAERITGFMPADEGRALYDTALEYLGDGVGV
TH_0085|M.thermoresistible__bu MTS--DHTTLPPAAARLFALADETTGFMPADEGRALYDAAMRFLGDGIGV
MSMEG_6482|M.smegmatis_MC2_155 MSH--ADTALPPRAERLFALAEQVTGFMPVDEGRALYDAAVRYLGGGVGV
MUL_1691|M.ulcerans_Agy99 ----------MLATIDKFAYEKSILVNVGDEKG-LLLDAAVRRADPALAL
* . : ::* * ::*:. . .:
MMAR_2922|M.marinum_M EIGTYCGKSTLLLGAAAQQCSSVLYTVDHHHGSEEHQPGWEYHDDSLVDE
MAV_2758|M.avium_104 EIGTYCGKSTLFLGAAAQQTGSVLYTVDHHHGSEEHQAGWEYHDASLVDE
Mflv_1102|M.gilvum_PYR-GCK EIGTYCGKSTVMLGAAAAQTGGLIYTVDHHHGSEEHQVGWEYHDASMVDP
Mvan_5715|M.vanbaalenii_PYR-1 EIGTYCGKSTVMLGAAAQQTGGLLYTVDHHHGSEEHQAGWEYHDASMVDP
TH_0085|M.thermoresistible__bu EIGTYCGKSTLLLGAAAQQTGGLLYTVDHHHGSEEHQPGWEYHDPSLVDP
MSMEG_6482|M.smegmatis_MC2_155 EIGTYCGKSTVMLGAAAQETGSVLYTVDHHHGSEEHQPGWEYHDASLVDP
MUL_1691|M.ulcerans_Agy99 ELGTYCGYGALRIARAAPQAK--VYSVELAEANAANAR-QIWAHAGVADR
*:***** .:: :. ** : :*:*: ... : : . .:.*
MMAR_2922|M.marinum_M ITGLFDTLPTFRRTLDSSGLDDHVVTVVGKSPIVARGWRSPLQLLFIDGG
MAV_2758|M.avium_104 VTGLFDTLPTFRRTLDAAGLDDHVVAVVGKSPIVARGWRSPLQLLFIDGG
Mflv_1102|M.gilvum_PYR-GCK VTGLFDTLPTLRHTLDAAGLDDHVVAIVGRSPVVARGWRTPLRFLFIDGG
Mvan_5715|M.vanbaalenii_PYR-1 VTGLFDTLPTLRHALDSAGLDDHVVAVVGRSPVVARGWRTPLRLLFIDGG
TH_0085|M.thermoresistible__bu VTGLFDTLPSLRHTLDAAGLDDHVVAVVGRSPVVARGWRTPLRLLFIDGG
MSMEG_6482|M.smegmatis_MC2_155 VTGLFDTLPKMRHTLDAAGLDDHVVAVVGRSPVVARGWRTPLRFLFIDGG
MUL_1691|M.ulcerans_Agy99 VTCVVGTIGDGGRTLDALATEHGFAP-------------GALDFVFVDHD
:* :..*: ::**: . :. ... .* ::*:* .
MMAR_2922|M.marinum_M HTEAAAAQDFEGWARWVSPGGALIIHDVFPNPADGGRAPYHIYRRAIDSG
MAV_2758|M.avium_104 HSETAANQDFDGWAKWVTPGGALAIHDVFPDPRDGGRPPYHIYCRAIDSG
Mflv_1102|M.gilvum_PYR-GCK HTEDAANRDFDGWARWVDVGGALVIHDVFPDPADGGRAPFLIYQRALKTG
Mvan_5715|M.vanbaalenii_PYR-1 HTEEAANRDFDGWARWVDVGGVLVIHDVFPDPADGGQAPFFIYKRALDCG
TH_0085|M.thermoresistible__bu HTEEAAQRDFEGWARWVDVGGALIIHDVFPDPADGGQAPYHIYRRALDTG
MSMEG_6482|M.smegmatis_MC2_155 HTEEAAQRDFDGWARWVQKGGALVIHDVFPNPDDGGQAPFHIYQRALATG
MUL_1691|M.ulcerans_Agy99 KKAYLDDLQSILDREWLHRG-SIVVADNVKVP---GAPEYHEYMRHHQGT
:. : .*: * : : * . * * . : * *
MMAR_2922|M.marinum_M QFQEISATGSLRLLERTSGGPGEQIEVSGRAC
MAV_2758|M.avium_104 QFREVSATGSLRVLERIGGRAGEPVEQD----
Mflv_1102|M.gilvum_PYR-GCK DFREVRATGSMRVLERVSGRPGELAA------
Mvan_5715|M.vanbaalenii_PYR-1 DFREVRAAGSMRVLERVSGQAGALGAA-----
TH_0085|M.thermoresistible__bu AFREVGATGSMRVLERTAGVAGMLTG------
MSMEG_6482|M.smegmatis_MC2_155 AFREVSATGSMRVLERISGEAGTLD-------
MUL_1691|M.ulcerans_Agy99 LWDTTEHKTHLEYQTLVSDLVLESDYLG----
: :. ..