For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSFTIPGLSDKKASDVADLLQKQLSTYNDLHLTLKHVHWNVVGPNFIGVHEMIDPQVELVRGYADEVAER IATLGKSPKGTPGAIIKDRTWDDYSVERDTVQAHLAALDLVYNGVIEDTRKSIEKLEDLDLVSQDLLIAH AGELEKFQWFVRAHLESAGGQLTHEGQSTEKGAADKARRKSA*
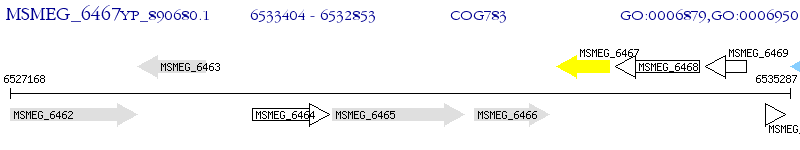
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6467 | - | - | 100% (183) | starvation-induced DNA protecting protein |
| M. smegmatis MC2 155 | MSMEG_3242 | - | 2e-11 | 31.16% (138) | starvation-inducible DNA-binding protein or fine tangled pili |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1112 | - | 2e-83 | 79.67% (182) | Ferritin, Dps family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3900c | - | 2e-84 | 80.22% (182) | hypothetical protein MAB_3900c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_5153 | - | 1e-81 | 78.02% (182) | starvation-induced DNA protecting protein |
| M. thermoresistible (build 8) | TH_2099 | - | 2e-80 | 78.41% (176) | starvation-induced DNA protecting protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5700 | - | 5e-86 | 81.87% (182) | Ferritin, Dps family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1112|M.gilvum_PYR-GCK ----------------MTTFTVPGLSDKQGAEVAELLQKALSRYNDLHLT
Mvan_5700|M.vanbaalenii_PYR-1 ----------------MTAFTVPGLTDKQGAEVAELLQKALSRYNDLHLT
MAB_3900c|M.abscessus_ATCC_199 ----------------MTEFTIPGLTDTQSRKIADLLQSQLSTYNDLHLT
MSMEG_6467|M.smegmatis_MC2_155 ----------------MTSFTIPGLSDKKASDVADLLQKQLSTYNDLHLT
MAV_5153|M.avium_104 ----------------MSEFTIPGLTDKQAARLTELLQKQLSTYNDLHLT
TH_2099|M.thermoresistible__bu VGYAWRRREKKELSMSKSHYTVPGMTDKDADQVIELLQKQLSCYNDLHLT
: :*:**::*... : :***. ** *******
Mflv_1112|M.gilvum_PYR-GCK LKHVHWNVVGPNFIGVHEMIDPQVELVRGYADEAAERIAALGTAPLGTPG
Mvan_5700|M.vanbaalenii_PYR-1 LKHVHWNVVGPNFIGVHEMIDPQVELVRGYADEVAERIAALGTSPLGTPG
MAB_3900c|M.abscessus_ATCC_199 LKHVHWNVVGPNFIGVHEMLDPQVELVRGYADAVAERIAALGRSPQGTPG
MSMEG_6467|M.smegmatis_MC2_155 LKHVHWNVVGPNFIGVHEMIDPQVELVRGYADEVAERIATLGKSPKGTPG
MAV_5153|M.avium_104 LKHIHWNVVGPNFIGVHEMIDPQVEAVRGFADDVAERIAALGASPQGTPG
TH_2099|M.thermoresistible__bu LKHVHWNVVGPNFIGVHEMIDPQVALIRGYADEVAERIATLGGSPKGTPG
***:***************:**** :**:** .*****:** :* ****
Mflv_1112|M.gilvum_PYR-GCK AIISDRTWDDYSVNRDTVQAHLAALDLVYTGVIEDTRKSIERLGELDPVS
Mvan_5700|M.vanbaalenii_PYR-1 AIINDRTWDDYSVARDTVQAHLAALDLVYTGVIEDTRKSIERLEELDLVS
MAB_3900c|M.abscessus_ATCC_199 AILKDRTWDDYSVGRDTVQAHLAALDLVYSGVIGDTRKAIEQLDELDLVS
MSMEG_6467|M.smegmatis_MC2_155 AIIKDRTWDDYSVERDTVQAHLAALDLVYNGVIEDTRKSIEKLEDLDLVS
MAV_5153|M.avium_104 AIIKDRSWDDYSVGRDTVQAHLAALDLVYNGVIEDIRQYIDETDELDQVT
TH_2099|M.thermoresistible__bu AILDDRTWDDYSIGRDCVQAHLSALDLVYNGVIEDTRKAIDETEELDPVT
**:.**:*****: ** *****:******.*** * *: *:. :** *:
Mflv_1112|M.gilvum_PYR-GCK EDMLIGHAAELEKFQWFVRAHLENSGGELVNEGATSEKAAASKAKNKS--
Mvan_5700|M.vanbaalenii_PYR-1 QDMLIGHAGELEKFQWFVRAHLENSGGELVNKGAATEKAAASKAKKNS--
MAB_3900c|M.abscessus_ATCC_199 QDLLIGHSGELEKFQWFVRAHLENSGGQLSNRDALTEKGAAARAKKKSPV
MSMEG_6467|M.smegmatis_MC2_155 QDLLIAHAGELEKFQWFVRAHLESAGGQLTHEGQSTEKGAADKARRKSA-
MAV_5153|M.avium_104 QDLLIGQAAQLEKFQWFVRAHLESAGGQLAHKGKSTERAAAQSARGKS--
TH_2099|M.thermoresistible__bu NDMLIGHAAEMEKFQWFVRAHLENAGGRLQHEGATTEKQAAAKAR-----
:*:**.::.::************.:**.* :.. :*: ** *:
Mflv_1112|M.gilvum_PYR-GCK -
Mvan_5700|M.vanbaalenii_PYR-1 -
MAB_3900c|M.abscessus_ATCC_199 K
MSMEG_6467|M.smegmatis_MC2_155 -
MAV_5153|M.avium_104 -
TH_2099|M.thermoresistible__bu -