For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RISLWLSVLVVAGLFGWGAWQRRWIADDGLIVLRTVRNLLAGNGPVFNAGERVEANTSTLWTYLVTLGAW IGGSVRLEYVVLAFALALSVAGVVLVMFGAGRLYAPSLQGRRALLLPAGALVYIAIPPARDFATSGLENG LVLAYLGLLWWMMVCWSQALRAYPADGGAPQDAPDNVPGRFFEGALAFVAGLSVLVRPELALIGGGALVM MLIAARGWRRRVLIVVAGGLLPVAYQIFRMGYYALIVPGTAVAKDATGSKWSQGLTYLANFNQPYLLWVP AVLLVLLGLVLVATRSRPWWLHRQVPRGYGWVRRTVQSPPAVVAFMVLSGLIQGIYWVRQGGDFMHGRVL LAPIFCVLAPVAVIPLVLPDGTRFTRETGYLLAAATSVLWVAVVGWSVWAANSPGMGADATRVTYTGIVD ERRFYSQATGHAHPLTAADYLDYPRMRAVLTALENTPDGALLLPSGNYDVWDVVPAIPPPPDMTPEQRKA YKAPHTVFFTNMGMLGMNVGLDVRVIDQIGLTNPLAQHTQRLTDGRIGHDKNLFPDWAVAEGPFLKEHPF VPAYLDEDWIAQAQVALTCPATDAVLNSVRGELTPRRFLSNIVHSYEFTKYRIDRVPLYELRRCGLEPPE PKNPPYTGMPATGP*
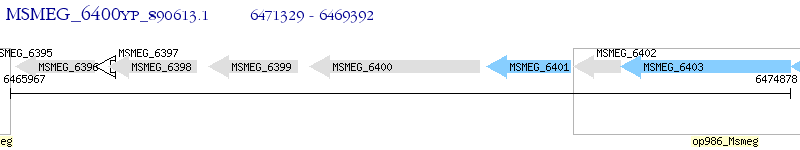
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6400 | - | - | 100% (645) | transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3835c | - | 0.0 | 72.01% (636) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1163 | - | 0.0 | 74.73% (645) | hypothetical protein Mflv_1163 |
| M. tuberculosis H37Rv | Rv3805c | - | 0.0 | 71.86% (636) | transmembrane protein |
| M. leprae Br4923 | MLBr_00096 | - | 0.0 | 69.58% (641) | hypothetical protein MLBr_00096 |
| M. abscessus ATCC 19977 | MAB_0174 | - | 0.0 | 66.11% (658) | hypothetical protein MAB_0174 |
| M. marinum M | MMAR_5369 | - | 0.0 | 72.37% (637) | transmembrane protein |
| M. avium 104 | MAV_0212 | - | 0.0 | 69.65% (636) | hypothetical protein MAV_0212 |
| M. thermoresistible (build 8) | TH_2080 | - | 0.0 | 75.49% (657) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4989 | - | 0.0 | 72.48% (636) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_5646 | - | 0.0 | 74.57% (645) | hypothetical protein Mvan_5646 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6400|M.smegmatis_MC2_155 ------------------------------------MRISLWLSVLVVAG
TH_2080|M.thermoresistible__bu VQPRSSVDRAAVAAAR-------RLAAWPPYPYSTSARISLWAGVLVVAG
Mflv_1163|M.gilvum_PYR-GCK MPQSSSADTV------------------------VRVRISLWLSVTAVAL
Mvan_5646|M.vanbaalenii_PYR-1 MPQSSSADTLSAAPGRVTAAKAGRLARWPAIPHDISVRVSLWLGVAVVAG
Mb3835c|M.bovis_AF2122/97 -----------------------------------MVRVSLWLSVTAVAV
Rv3805c|M.tuberculosis_H37Rv -----------------------------------MVRVSLWLSVTAVAV
MMAR_5369|M.marinum_M ---------------------MLRPIRWPAFPYDPWVRISLWIGVAIIAL
MUL_4989|M.ulcerans_Agy99 ---------------------MLRPIRWPAFPYDPWVRISLWIGVAIIAL
MAV_0212|M.avium_104 -MPSVSPELQALKARIMGSRPVMRRPGWPVFPYTTMVRSSLWIGVAVVAV
MLBr_00096|M.leprae_Br4923 ----------------MRRRMVSRQVGWPLFPYHIVVRVSLWASVLLVAA
MAB_0174|M.abscessus_ATCC_1997 -----------------------------MGPSNGLTRFTVWVSVLTVTV
* ::* .* ::
MSMEG_6400|M.smegmatis_MC2_155 LFGWGAWQRRWIADDGLIVLRTVRNLLAGNGPVFNAGERVEANTSTLWTY
TH_2080|M.thermoresistible__bu LFGYGAWQRRWIADDGLIVLRTVRNLLAGNGPVFNAGERVEANTSTVWTY
Mflv_1163|M.gilvum_PYR-GCK LFGWGAWQRRWIADDGLIVLRTTRNMLAGNGPVFNAGERVEANTSTAWSY
Mvan_5646|M.vanbaalenii_PYR-1 LFGWGAWQRRWIADDGLIVLRTTRNMLAGNGPVFNAGERVEANTSTVWSY
Mb3835c|M.bovis_AF2122/97 LFGWGSWQRRWIADDGLIVLRTVRNLLAGNGPVFNQGERVEANTSTAWTY
Rv3805c|M.tuberculosis_H37Rv LFGWGSWQRRWIADDGLIVLRTVRNLLAGNGPVFNQGERVEANTSTAWTY
MMAR_5369|M.marinum_M LFGWGAWQRRWIADDGLIVLRTVRNLLAGNGPVFNMGERVEANTSTVWTY
MUL_4989|M.ulcerans_Agy99 LFGWGAWQRRWIADDGLIVLRTVRNLLAGNGPVFNMGERVEANTSTVWTY
MAV_0212|M.avium_104 LFAWGGWQRRWIADDGLIVLRTVRNLLAGNGPVFNQGERVEANTSTAWTY
MLBr_00096|M.leprae_Br4923 LFGWGAWQRRWIADDGLIVLRTVRNLLAGNGPVFNQGERVEANTSTVWTY
MAB_0174|M.abscessus_ATCC_1997 LFGWGAWQRRWIADDGLIVLRTVRNLLAGNGPVFNKGERVEANTSTLWTY
**.:*.****************.**:********* ********** *:*
MSMEG_6400|M.smegmatis_MC2_155 LVTLGAWIGGSVRLEYVVLAFALALSVAGVVLVMFGAGRLYAPSLQGRRA
TH_2080|M.thermoresistible__bu LNYLGALLAGPVRLEYVILVLAFGLSVLGVILAMLGAGRLYAPGLQGRPA
Mflv_1163|M.gilvum_PYR-GCK LMYFGGLVAGPARLEYVALTFALILSVLGVVLVMLGTARLYAPVLRGRKA
Mvan_5646|M.vanbaalenii_PYR-1 LMYLGGLVAGPARLEYVALTLALVLSVLGVVLVMLGTARLYEPTLKGRRA
Mb3835c|M.bovis_AF2122/97 LLYVGGWVGGPMRLEYVALALAMVLSLLGMVLLMLGTGRLYAPSLRGRRA
Rv3805c|M.tuberculosis_H37Rv LLYVGGWVGGPMRLEYVALALAMVLSLLGMVLLMLGTGRLYAPSLRGRRA
MMAR_5369|M.marinum_M LLYAASWVGGPMRLEYVALAVALTLSVLGVVLLMLGAGRLYAPSLRGRRA
MUL_4989|M.ulcerans_Agy99 LLYAASWVGGPMRLEYVALAVALTLSVLGVVLVMLGAGRLYAPSLRGRRA
MAV_0212|M.avium_104 LMYAGSWVGGPLRMEYVALALALTLSVLGVALLMLGAARLYAPSLRGRRA
MLBr_00096|M.leprae_Br4923 LLYAGSWVGGPMRFEYVALAAALVLSVLGMVLLMLGTGRLYAPSLQGRQA
MAB_0174|M.abscessus_ATCC_1997 LTYLGGWAGGGMRLEYVALTLSLVLSLVGVALAILGTARLYAPLLAGRAA
* .. .* *:*** *. :: **: *: * ::*:.*** * * ** *
MSMEG_6400|M.smegmatis_MC2_155 LLLPAGALVYIAIPPARDFATSGLENGLVLAYLGLLWWMMVCWSQALRAY
TH_2080|M.thermoresistible__bu LLLPAGILVYIAVPPARDFATSGLENGLVLAYLALLWWMMVCWAQAPRRR
Mflv_1163|M.gilvum_PYR-GCK LLLPAGALVYIAVPPARDFATSGLENGLVLAYLGLLWWMMVAWSQARR--
Mvan_5646|M.vanbaalenii_PYR-1 LLLPAGALVYIAVPPARDFATSGLENGLVLAYLGLLWWMMVRWSQGLRGR
Mb3835c|M.bovis_AF2122/97 IMLPAGALVYIAVPPARDFATSGLESGLVLAYLGLLWWMMVCWSQPLRAR
Rv3805c|M.tuberculosis_H37Rv IMLPAGALVYIAVPPARDFATSGLESGLVLAYLGLLWWMMVCWSQPLRAR
MMAR_5369|M.marinum_M IMLPAGALVYIAVPPARDFATSGLESGLVLAYLGLLWWMMVCWSQPLRGR
MUL_4989|M.ulcerans_Agy99 IMLPAGALVYIAVPPARDFATSGLESGLVLAYLGLLWWMMVCWSQPLRGR
MAV_0212|M.avium_104 IMLPAGALVYIALPPSRDFATSGLESGLALAYLGLLWWMMVCWAQPVRNR
MLBr_00096|M.leprae_Br4923 IMLPAGALVYVALPPARDFATSGLESGLVLTYLGLLWWMMVCWAQPLRNR
MAB_0174|M.abscessus_ATCC_1997 VMVPAGMLVYIAIPPARDFATSGLENGLVLAYLGGLWLMMVKWAQAVRTP
:::*** ***:*:**:*********.**.*:**. ** *** *:* *
MSMEG_6400|M.smegmatis_MC2_155 PADGG-APQDAPDN------------VPGRFFEGALAFVAGLSVLVRPEL
TH_2080|M.thermoresistible__bu PAGAGTTPRTAAANP------TDTGETVGAGFHAALAFVAGMSVLVRPEL
Mflv_1163|M.gilvum_PYR-GCK ETTRT--------------------------FDAALAAVAGFSVLVRPEL
Mvan_5646|M.vanbaalenii_PYR-1 PTSRS--------------------------FDVTLAVVAGLSVLVRPEL
Mb3835c|M.bovis_AF2122/97 PDSQM--------------------------FLGALAFVAGCSVLVRPEF
Rv3805c|M.tuberculosis_H37Rv PDSQM--------------------------FLGALAFVAGCSVLVRPEF
MMAR_5369|M.marinum_M SDHPV--------------------------FIGLLAFVAGCSVLVRPEL
MUL_4989|M.ulcerans_Agy99 SDNPV--------------------------LIGLLAFVAGCSVLVRPEL
MAV_0212|M.avium_104 SDRRE--------------------------FAGALAFVAGFSVLVRPEL
MLBr_00096|M.leprae_Br4923 SQSRR--------------------------FIGALAFVAGCSVLVRPEL
MAB_0174|M.abscessus_ATCC_1997 VTLDRRGGPHQPVDPRIVHAQRDQRDVLSRRFTVGLAFLAGLSVLIRPEL
: ** :** ***:***:
MSMEG_6400|M.smegmatis_MC2_155 ALIGGGALVMMLIAARGWRRRVLIVVAGGLLPVAYQIFRMGYYALIVPGT
TH_2080|M.thermoresistible__bu ALIGGLALIMILIAARTRRRRLLIVVAGGLVPVAYQIFRMGYYALVVPGT
Mflv_1163|M.gilvum_PYR-GCK ALIGGLALVMMLIAAGGWRRRALIFVAGGLLPVAYQIFRMGYYGLLFPGT
Mvan_5646|M.vanbaalenii_PYR-1 ALIGGLALVMMLIAVDQWRTRALIVVAGGLLPVGYQIFRMGYYGLLFPGT
Mb3835c|M.bovis_AF2122/97 ALIGGLALIMMLIAARTWRRRVLIVLAGGFLPVAYQIFRMGYYGLLVPST
Rv3805c|M.tuberculosis_H37Rv ALIGGLALIMMLIAARTWRRRVLIVLAGGFLPVAYQIFRMGYYGLLVPST
MMAR_5369|M.marinum_M ALIGGLALIMMLIAARSWRRRGLIVLAGGLLPVAYQIFRMGYYGLLVPGT
MUL_4989|M.ulcerans_Agy99 ALIGGLALIMMLIAARSWRRRGLIVLAGGLLPVAYQIFRMGYYGLLVPGT
MAV_0212|M.avium_104 ALMGGLALIMMLVAARTWRRRALIVAAGGALPVAYEIFRMGYYGLLVPGT
MLBr_00096|M.leprae_Br4923 ALMGGSALIMLMIAARTCWLRALIVVAGGSLPVAYQLFRMGYYGLLVPGT
MAB_0174|M.abscessus_ATCC_1997 ALIGGGFLVMMLVAARGFMSRIWIVVAGGALPVLYQIFRMGYYGLLVPST
**:** *:*:::*. * *. *** :** *::******.*:.*.*
MSMEG_6400|M.smegmatis_MC2_155 AVAKDATGSKWSQGLTYLANFNQPYLLWVPAVLLVLLGLVLVATRSRPWW
TH_2080|M.thermoresistible__bu AVAKEAGGAKWSQGLAYLTNFTSPYLLWLPMLLVAALGAVLLATRLRPWR
Mflv_1163|M.gilvum_PYR-GCK ALAKDATGAKWQQGFTYLANFNQPYLLWAPAVLLIGLAVMVMLLCGRPPR
Mvan_5646|M.vanbaalenii_PYR-1 ALAKDATGAKWEQGLVYLANFNQPYLLWAPAVLLIGLAIMVRLLCGRPAG
Mb3835c|M.bovis_AF2122/97 ALAKDAAGDKWSQGMIYVSNFNRPYALWVPLVLSVPLGLLLMTARRRPSF
Rv3805c|M.tuberculosis_H37Rv ALAKDAAGDKWSQGMIYVSNFNRPYALWVPLVLSVPLGLLLMTARRRPSF
MMAR_5369|M.marinum_M ALAKDAAGDKWSQGMVYLTNFNRPYALWLPLVLVVPLGVVVMFARRRPAF
MUL_4989|M.ulcerans_Agy99 ALAKDAAGDKWSQGMVYLTNFNRPYALWLPLVLVVPLGVVVMFARRRPAF
MAV_0212|M.avium_104 ALAKDAAGDKWSQGMTYLSNFDAPYLVWVPAALLVLLGVLLAAARRRPSF
MLBr_00096|M.leprae_Br4923 ALAKDAAGDKWSQGIIYLSNFNQPYVLWVPLVLLVLLGLLLMLIHRWPSF
MAB_0174|M.abscessus_ATCC_1997 AIAKDASGSKWGQGFVYLQNMNSPYLIWIPAVLLIALGVAAYQARRGQWW
*:**:* * ** **: *: *: ** :* * * *.
MSMEG_6400|M.smegmatis_MC2_155 LHRQVPRGYGWVRRTVQSPPAVVAFMVLSGLIQGIYWVRQGGDFMHGRVL
TH_2080|M.thermoresistible__bu RH-AVPRNLGWVARTLQSPPVVVAFFVAGGVLQAIYWIRQGGDFMHGRVL
Mflv_1163|M.gilvum_PYR-GCK VDRRP--GSGRLARLVQSPSAVVVFMLLSGLLQAVYWIRQGGDFMHGRVL
Mvan_5646|M.vanbaalenii_PYR-1 VDRRASAGSGWLARTVRSPSAVVLFMLVSGALQAIYWIRQGGDFMHGRVL
Mb3835c|M.bovis_AF2122/97 LRPVLAPDYGRVARAVQSPPAVVAFIVGSGVLQALYWVRQGGDFMHGRVL
Rv3805c|M.tuberculosis_H37Rv LRPVLAPDYGRVARAVQSPPAVVAFIVGSGVLQALYWIRQGGDFMHGRVL
MMAR_5369|M.marinum_M LRPVLAPDYGRFARAVQSPTAVVVFMVGSGVLQALYWIRQGGDFMHGRVL
MUL_4989|M.ulcerans_Agy99 LRPVLAPGYGRFARAVQSPTAVVVFMVGSGVLQALYWIRQGGDFMHGRVL
MAV_0212|M.avium_104 LRPPLAPNYGRVARAVHSPPAVVAWVLVSGLLQALYWIRQGGDFMHARVL
MLBr_00096|M.leprae_Br4923 MHPLETPDSGRVARAVQSPPAVVVFVVFSGLLQAFYWIRQGGDFMHGRVL
MAB_0174|M.abscessus_ATCC_1997 VRQVAAPGYGWLARLVQNPTAVVVFMLVSGFVQGVYWIRQGGDFMHARVL
. * . * ::.*..** :.: .* :*..**:********.***
MSMEG_6400|M.smegmatis_MC2_155 LAPIFCVLAPVAVIPLVLPDGTRFT-----RETGYLLAAATSVLWVAVVG
TH_2080|M.thermoresistible__bu LTPLFLILLPVAVVPLRLPGGTAAADSDERRRHQYVLAGATGVLWVAIAG
Mflv_1163|M.gilvum_PYR-GCK LTPLFCLLAPVSVIPLLLPDGRRMA-----RGAGYLYAGATSLLWLAVAG
Mvan_5646|M.vanbaalenii_PYR-1 LTPLFCLLAPVSVVPLMLPDSRRMA-----RGAGYLYAGATSVLWLAVAG
Mb3835c|M.bovis_AF2122/97 LAPLFCLLAPVGVIPILLPDGKDFS-----RETGRWLVGALSGLWLGIAG
Rv3805c|M.tuberculosis_H37Rv LAPLFCLLAPVGVIPILLPDGKDFS-----RETGRWLVGALSGLWLGIAG
MMAR_5369|M.marinum_M LAPLFCVLAPVAVIPLVVPDGKDYS-----RETGYWVAGAVSVLWLGVAG
MUL_4989|M.ulcerans_Agy99 LAPLFCVLAPVAVIPLVVPDGKDYS-----RETGYWVAGAVSVLWLGVAG
MAV_0212|M.avium_104 LAPLFCLLAPVAVVPVMVPDGQDYS-----RETGNWVAGAACALWLGIAG
MLBr_00096|M.leprae_Br4923 LAPLFCLLAPVVVIPVVISEGADFS-----RQTGNWLAGVTSLLWLGVAG
MAB_0174|M.abscessus_ATCC_1997 LTPVFCMLLPISVVPLAAPDSAAFT-----PKKARLLTAATIGLFAGIAG
*:*:* :* *: *:*: . . : ... *: .:.*
MSMEG_6400|M.smegmatis_MC2_155 WSVWAANSPGMGADATRVTYTGIVDERRFYSQATGHAHPLTAADYLDYPR
TH_2080|M.thermoresistible__bu WALWAANSPGMGPDATKVTYSGIVDERRFYAQATGHAHPLTAADYLDYPR
Mflv_1163|M.gilvum_PYR-GCK WSLWAAHSPGMGADATRVTYTGIVDERRFYAQATGHAHPLTAADYLDYPR
Mvan_5646|M.vanbaalenii_PYR-1 WALWAANSPGMGADATRVTYTGIVDERRFYSQATGHAHPLTAADYLDYPR
Mb3835c|M.bovis_AF2122/97 WSLWAANSPGMGDDATRVTYSGIVDERRFYAQATGHAHPLTAADYLDYPR
Rv3805c|M.tuberculosis_H37Rv WSLWAANSPGMGDDATRVTYSGIVDERRFYAQATGHAHPLTAADYLDYPR
MMAR_5369|M.marinum_M WSLWAANSPGMGDDATHVSYSGIVDERRFYAQATGHAHPLTAADYLDYPR
MUL_4989|M.ulcerans_Agy99 WSLWAANSPGMGDDATHVSYSGIVDERRFYAQATGHAHPLTAADYLDYPR
MAV_0212|M.avium_104 WALWAANSPGMGDDATHVTYSGIVDERRFYAQATGHAHPLTAADYLGYPR
MLBr_00096|M.leprae_Br4923 WSLWAANSPGMGDDATNVSYSGIVDERRFYAQATGHAHPLTAADYLGYPR
MAB_0174|M.abscessus_ATCC_1997 WSVWVANSPGMAGDGTKVTYSGIVDERRFYAQATGVAHPLTAADYLNYPR
*::*.*:****. *.*.*:*:*********:**** **********.***
MSMEG_6400|M.smegmatis_MC2_155 MRAVLTALENTPDGALLLPSGNYDVWDVVPAIPPPP---DMTPEQRKAYK
TH_2080|M.thermoresistible__bu MRAVLVTLNNTPEGALLLPSGNYDQWDVVPAIPPPP---DAPPEARDPDN
Mflv_1163|M.gilvum_PYR-GCK MRAVVTAIENTPDGALLLPSGNYDMWDVVPAYPPPP---DAP----PDYR
Mvan_5646|M.vanbaalenii_PYR-1 MRAVLTAIENTPDGALLLPSGNYDMWDVVPAYPPPP---DAPRGFGSEYN
Mb3835c|M.bovis_AF2122/97 MAAVLTALNNTPEGALLLPSGNYNQWDLVPMIRPSS---GTAPGGKPAPK
Rv3805c|M.tuberculosis_H37Rv MAAVLTALNNTPEGALLLPSGNYNQWDLVPMIRPSS---GTAPGGKPAPK
MMAR_5369|M.marinum_M MAAILTALNNTPEGALLLPSGNYTQWDLVPMIKP-----GSAPGIPASEK
MUL_4989|M.ulcerans_Agy99 MAAILTALNNTPDGALLLPSGNYTQWDLVPMIKP-----GSAPGIPASEK
MAV_0212|M.avium_104 MAAILTALNNTPEGALLLPSGNYTQWDLVPMAQPPP---GTTPDKFP-QK
MLBr_00096|M.leprae_Br4923 MAAVLVALNNTPDGALLLPSGNYIKWDLVPMIQLSPSSPGSPPDSLVSQK
MAB_0174|M.abscessus_ATCC_1997 MRAVLVAIDNTPDGALLLPSGNYDQWDVAPAIPPPP-------PIPHGYR
* *::.:::***:********** **:.* .
MSMEG_6400|M.smegmatis_MC2_155 APHTVFFTNMGMLGMNVGLDVRVIDQIGLTNPLAQHTQRLTDGRIGHDKN
TH_2080|M.thermoresistible__bu APHTVFFTNLGMLGMNVGLDVRVIDQIGLANPLAAHTERVEDGRIGHDKN
Mflv_1163|M.gilvum_PYR-GCK GPHTVFFTNMGMLGMNVGLDVRVIDQIGLANPLAAHTARIEDGRIGHDKN
Mvan_5646|M.vanbaalenii_PYR-1 GPHTVFFTNLGMLGMNVGLDVRVIDQIGLANPLAAHTARIEDGRIGHDKN
Mb3835c|M.bovis_AF2122/97 PQHAVFFTNMGMLGMNVGLDVRVIDQIGLVNPLAAHTERLKHARIGHDKN
Rv3805c|M.tuberculosis_H37Rv PQHAVFFTNMGMLGMNVGLDVRVIDQIGLVNPLAAHTERLKHARIGHDKN
MMAR_5369|M.marinum_M PQHTVFFTNLGMVGMNVGLDVRVLDQIGLANPLAAHTERLEHGRIGHDKN
MUL_4989|M.ulcerans_Agy99 PQHTVFFTNLGMVGMNVGLDVRVLDQIGLANPLAAHTERLEHGRIGHDKN
MAV_0212|M.avium_104 PQHTVFFTNLGMVGMNVGLDVRVIDQIGLANPLAQHTERLKHGRIGHDKN
MLBr_00096|M.leprae_Br4923 PQHTVFFTNLGMLGMNVGLDVRVIDQIGLANPLAQHTERLQHGRIGHDKN
MAB_0174|M.abscessus_ATCC_1997 GPHAVLFTNLGMLGMNLGLDVRIVDQIGLANPLAAHTARITDGRIGHDKN
*:*:***:**:***:*****::*****.**** ** *: ..*******
MSMEG_6400|M.smegmatis_MC2_155 LFPDWAVAEGPFLKEHPFVPAYLDEDWIAQAQVALTCPATDAVLNSVRGE
TH_2080|M.thermoresistible__bu LFPDWAVAEGPFLKTYPWIPSYLDEDWVAQAEAALACPATDAKLNSVRGR
Mflv_1163|M.gilvum_PYR-GCK LFPDWAVAEGPFFKVEPYIPDYLDEDWIRQAEVALTCPETERMLDAIRAP
Mvan_5646|M.vanbaalenii_PYR-1 LFPDWAVAEGPFFKEPPYIPTYLDEDWIRQAEAALKCPETEAVLNAIRAP
Mb3835c|M.bovis_AF2122/97 LFPDWVIADGPWVKWYPGIPGYIDQQWVTQAEAALQCPATRAVLNSVRAP
Rv3805c|M.tuberculosis_H37Rv LFPDWVIADGPWVKWYPGIPGYIDQQWVTQAEAALQCPATRAVLNSVRAP
MMAR_5369|M.marinum_M LFPDWVIADGPWVKWYPGVPAYLDQQWVAQAEAALRCPATQAVLNSVRAP
MUL_4989|M.ulcerans_Agy99 LFPDWVIADGPWVKWYPGVPAYLDQQWVAQAEAALRCPATQAVLNSVRAP
MAV_0212|M.avium_104 LFPDWVIADGPWVKVYPGIPGYLDAQWVAEAVSALQCPDTRAVLASVRAP
MLBr_00096|M.leprae_Br4923 LFPDWVIADGPWVKWYPGIPGYLDQAWIAQAVAALQCSGTQAVLSSVRAP
MAB_0174|M.abscessus_ATCC_1997 LFPDWMIADGPWLKRYPYIPRYIDQDWVAEAVEALKCPQTDAMLSAVRKP
***** :*:**:.* * :* *:* *: :* ** *. * * ::*
MSMEG_6400|M.smegmatis_MC2_155 LTPRRFLSNIVHSYEFTKYRIDRVPLYELRRCGLEPPEPKNPPYTGMPAT
TH_2080|M.thermoresistible__bu LTPRLFVSNLVHALEFTRYRIDRVPRYELLRCGLEMPPPKQPPYTGLPAT
Mflv_1163|M.gilvum_PYR-GCK MGPRRFLSNLVNAAEYTRYRLDRVPLYELHRCGLPIPEPINPQYTGMPPT
Mvan_5646|M.vanbaalenii_PYR-1 MGPRRFLSNLLNAAEFTRYRIDRVPMYELERCGLPLPEPVNPPYTGLPPT
Mb3835c|M.bovis_AF2122/97 ITLHRFLSNVLHSYEFTRYRIDRVPRYELVRCGLDVPDGPGPPPRE----
Rv3805c|M.tuberculosis_H37Rv ITLHRFLSNVLHSYEFTRYRIDRVPRYELVRCGLDVPDGPGPPPRE----
MMAR_5369|M.marinum_M MSLHRFVSNVVHSFEFTRYRINRVPLYELVRCGLEVPDVPPAPPRE----
MUL_4989|M.ulcerans_Agy99 MSLHRFVSNVVHSFEFTRYRINRVPLYELVRCGLEVPDVPPAPSRE----
MAV_0212|M.avium_104 MTPHRFLSNVLHSFQFTGYRIDRVPRYELARCGLPVPEEAPPPPRE----
MLBr_00096|M.leprae_Br4923 MALHRFISNLLNSFEFTRYRFDRVPLYELVRCGLPVPDVLPATPPE----
MAB_0174|M.abscessus_ATCC_1997 LSPRLFVSNMLHSYEFTTYRIDRVPRFELARCGLPMPKLDTPSYTGLPAT
: : *:**:::: ::* **::*** :** **** * .
MSMEG_6400|M.smegmatis_MC2_155 GP
TH_2080|M.thermoresistible__bu GP
Mflv_1163|M.gilvum_PYR-GCK GP
Mvan_5646|M.vanbaalenii_PYR-1 GP
Mb3835c|M.bovis_AF2122/97 --
Rv3805c|M.tuberculosis_H37Rv --
MMAR_5369|M.marinum_M --
MUL_4989|M.ulcerans_Agy99 --
MAV_0212|M.avium_104 --
MLBr_00096|M.leprae_Br4923 --
MAB_0174|M.abscessus_ATCC_1997 GP