For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DQPYFTVTAELEGRLGRVGTIHTPHGDIHTPAFIPVGTAATVKSVLPETMKQLGAQAILANAYHLYLQPG PDIVAEAGGLGAFMNWPGPTFTDSGGFQVLSLGVGFKKVLAMDTNRVQADDIIAEGKERLAHVDDDGVTF RSHLNGSLHRFTPEVSIGIQYQLGADIIFAFDELTTLVNTRAYQERSVQRTHEWAERCLAEHHRLQNQAP ERPRQALFGVVQGAQYEDLRRKAARGLATIGEQAGPAEGLAFDGYGIGGALEKQNLATIVGWVSSELPKD KPRHMLGISEPDDLFAAVAAGADTFDCVSPSRVARNAAIYTSTGRYNITGARYKRDFTPLDPECDCYTCA NYSRAYIRHLFKAKEMLSATLCTIHNERFVVRLVDRIRAAMLAGEFDALRDEVLGRYYGTSVR*
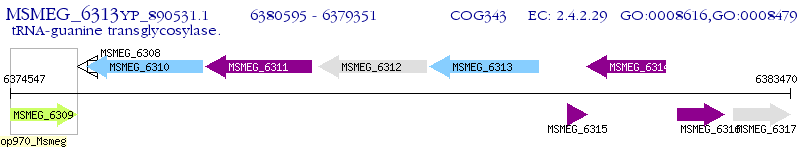
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6313 | tgt | - | 100% (414) | queuine tRNA-ribosyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1228 | - | 0.0 | 82.84% (402) | queuine tRNA-ribosyltransferase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0275 | - | 0.0 | 76.35% (406) | queuine tRNA-ribosyltransferase (TGT) |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1651 | tgt | 0.0 | 83.05% (407) | queuine tRNA-ribosyltransferase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5580 | - | 0.0 | 82.09% (402) | queuine tRNA-ribosyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1228|M.gilvum_PYR-GCK -----------MTAELPGRLGRTGVIRTPHGEIRTPAFIAVGTQATVKAV
Mvan_5580|M.vanbaalenii_PYR-1 -----------MDVELPGRLGRTGTIRTPHGDIQTPAFIAVGTQATVKAV
MSMEG_6313|M.smegmatis_MC2_155 ----MDQPYFTVTAELEGRLGRVGTIHTPHGDIHTPAFIPVGTAATVKSV
TH_1651|M.thermoresistible__bu VTSDSSGPFFSVGTELPGRLGRTGVIRTPHGEIHTPAFIPVGTQATVKAV
MAB_0275|M.abscessus_ATCC_1997 ----MTDPYFSIDATLSGTAGRAGVIHTPHGDIQTPAFIAVGTKATVKAV
: . * * **.*.*:****:*:*****.*** ****:*
Mflv_1228|M.gilvum_PYR-GCK LPETMKQVGAQAVLANAYHLYLQPGPDIVDEAGGLGAFMNWPGPTFTDSG
Mvan_5580|M.vanbaalenii_PYR-1 LPETMKEIGAQAVLANAYHLYLQPGADVVDEAGGLGAFMNWPGPTFTDSG
MSMEG_6313|M.smegmatis_MC2_155 LPETMKQLGAQAILANAYHLYLQPGPDIVAEAGGLGAFMNWPGPTFTDSG
TH_1651|M.thermoresistible__bu LPETMKELGAQAVLANAYHLYLQPGPDIVAEAGGLGAFMNWPGPTFTDSG
MAB_0275|M.abscessus_ATCC_1997 LPETMAALGAQAVLANAYHLYLQPGPDIVDEAGGLGAFMNWPGPTFTDSG
***** :****:************.*:* ********************
Mflv_1228|M.gilvum_PYR-GCK GFQVMSLGVGFKKVLAMDTERVRADDIIAEGKERLAHVDDDGVTFRSHLD
Mvan_5580|M.vanbaalenii_PYR-1 GFQVMSLGVGFKKVLAMDTSRVQADDIIAEGKSRLAHVDDDGVTFRSHLN
MSMEG_6313|M.smegmatis_MC2_155 GFQVLSLGVGFKKVLAMDTNRVQADDIIAEGKERLAHVDDDGVTFRSHLN
TH_1651|M.thermoresistible__bu GFQVLSLGAGFRKVLAMDANRVQADDVIAEGKERLAHVDDDGVTFRSHLD
MAB_0275|M.abscessus_ATCC_1997 GFQVLSLGSGVRKVMAMDVNRARADDVTVAGKDKLAHVDDDGVTFTSHLD
****:*** *.:**:***..*.:***: . **.:*********** ***:
Mflv_1228|M.gilvum_PYR-GCK GSSHRFTPEVSIGIQHQLGADIIFAFDELTTLVNTREYQESSVQRTHEWA
Mvan_5580|M.vanbaalenii_PYR-1 GSSHRFTPEVSIGIQHQLGADIIFAFDELTTLVNTRGYQERSVQRTHDWA
MSMEG_6313|M.smegmatis_MC2_155 GSLHRFTPEVSIGIQYQLGADIIFAFDELTTLVNTRAYQERSVQRTHEWA
TH_1651|M.thermoresistible__bu GSTHRFTPEKSIGIQRRLGADIIFAFDELTTLVNTRGYQERSVRRTHEWA
MAB_0275|M.abscessus_ATCC_1997 GSAHRFTPEVSMQIQHQLGADIMFAFDELTTLINTREYQEDSVQRTHEWA
** ****** *: ** :*****:*********:*** *** **:***:**
Mflv_1228|M.gilvum_PYR-GCK VRCLVEHRRLSDVRSHTPQQALFGVVQGAQYEDLRRQAARGLTGIVD---
Mvan_5580|M.vanbaalenii_PYR-1 VRCLAEHRRLSRVRSHTPPQALFGVVQGAQYEDLRRQAARGLTQIVD---
MSMEG_6313|M.smegmatis_MC2_155 ERCLAEHHRLQNQAPERPRQALFGVVQGAQYEDLRRKAARGLATIGEQAG
TH_1651|M.thermoresistible__bu VRCLVEHHRLRAAHPELPRQALFGVVQGAQYEDLRRQAARGLTEIGAGPG
MAB_0275|M.abscessus_ATCC_1997 QRCLDEHEKLTRERADKPYQALFGVVQGAQYEDLRRQAARGLESLAG---
*** **.:* .. * *****************:***** :
Mflv_1228|M.gilvum_PYR-GCK -EHGRGFDGYGIGGALEKQNLATIVGWVSSELPDDKPRHLLGISEPDDLF
Mvan_5580|M.vanbaalenii_PYR-1 -EEGRGFDGYGIGGALEKQNLATIVGWVSSELPDDKPRHLLGISEPDDLF
MSMEG_6313|M.smegmatis_MC2_155 PAEGLAFDGYGIGGALEKQNLATIVGWVSSELPKDKPRHMLGISEPDDLF
TH_1651|M.thermoresistible__bu PAEGLGFDGYGIGGALEKQNLATIVGWVSSELPPDKPRHLLGISEPDDLF
MAB_0275|M.abscessus_ATCC_1997 -PSGRGFDGYGIGGALEKQNLGTIVGWVTSELPEHKPRHLLGISEPDDLF
* .***************.******:**** .****:**********
Mflv_1228|M.gilvum_PYR-GCK AAVEAGADTFDCVSPSRVARNAAVYSADGRYNITGARYRRDFTPIDAECD
Mvan_5580|M.vanbaalenii_PYR-1 AAIEAGADTFDCVSPSRVARNAAVYSADGRFNITGSRYRRDFTPIDAECD
MSMEG_6313|M.smegmatis_MC2_155 AAVAAGADTFDCVSPSRVARNAAIYTSTGRYNITGARYKRDFTPLDPECD
TH_1651|M.thermoresistible__bu AAVAAGADTFDCVSPSRVARNAAVYSSTGRFNVTNARFRRDFNPIDDGCD
MAB_0275|M.abscessus_ATCC_1997 AAVAAGADTFDCVSPSRVARNAAVYTRDGRVNITGARYRRDFTPIDAECD
**: *******************:*: ** *:*.:*::***.*:* **
Mflv_1228|M.gilvum_PYR-GCK CYTCANYTRAYLHHLFKAKELLAATLCTIHNERFIIRLVDDIRASIPAGR
Mvan_5580|M.vanbaalenii_PYR-1 CYTCTHYTRAYLHHLFKAKEMLSATLCTIHNERFVIRLVDDIRVAIREGR
MSMEG_6313|M.smegmatis_MC2_155 CYTCANYSRAYIRHLFKAKEMLSATLCTIHNERFVVRLVDRIRAAMLAGE
TH_1651|M.thermoresistible__bu CYTCAHYTRAYLHHLFKARELLSATLATIHNQRFIIRLVDRIRESIVAGE
MAB_0275|M.abscessus_ATCC_1997 CYTCAHYTRAYMHHLFKAKEILASTLATIHNERFTIGLVDRIRASIVEGC
****::*:***::*****:*:*::**.****:** : *** ** :: *
Mflv_1228|M.gilvum_PYR-GCK FDELREHVLGRYYAAKS-------
Mvan_5580|M.vanbaalenii_PYR-1 FDELREHVLGRYYAKA--------
MSMEG_6313|M.smegmatis_MC2_155 FDALRDEVLGRYYGTSVR------
TH_1651|M.thermoresistible__bu FDELREHTLGRYYGGRRRYGTMAT
MAB_0275|M.abscessus_ATCC_1997 FQELREDTLGRYYR----------
*: **:..*****