For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RILLVGAGGVGSAFCAIAARRNFFDQIVVCDYDESRARQAAEAVGDARFTSAQVDATSADAVADLVREHQ ITHVMNAVDPRFVMPIFNGALAGGADYLDMAMSLSRRHPDRPYELTGVKLGDEQFGAAEDWEAAGRLALV GIGVEPGLSDVFARYAADHLFSEIDELGTRDGSSLVVEGCDFAPSFSIWTTIEECLNPPVVWEADRGWFV TEPFSEPEVFDFPEGIGPVECVNVEHEEVLLMPRWVKCKRATFKYGLGSEFIDVLKTLHKLGLDRTDKVR VPGANGPVEVSPRDVVAACLPNPATLGPQMHGKTCAGLWVTGKGKDGAPRSTYLYHVVDNQWSMAEYGHQ CVVWQTAINPVVALELLASGMWSGAGVLGPEAFDAVPFLNLLKDYGSPWGIKEL*
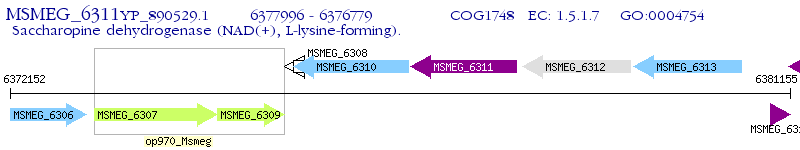
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6311 | lys1 | - | 100% (405) | saccharopine dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3325c | - | 1e-176 | 72.28% (404) | saccharopine dehydrogenase |
| M. marinum M | MMAR_3555 | - | 0.0 | 83.46% (405) | saccharopine dehydrogenase |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2788 | - | 0.0 | 82.96% (405) | saccharopine dehydrogenase |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3555|M.marinum_M MRILLVGAGGVGSAFCAIAARRSFFEHVVVCDYDEARASRAAQAVADARF
MUL_2788|M.ulcerans_Agy99 MRILLVGAGGVGSAFCAIAARRSFFEHVVVCDYDEARASRAAQAVADARF
MSMEG_6311|M.smegmatis_MC2_155 MRILLVGAGGVGSAFCAIAARRNFFDQIVVCDYDESRARQAAEAVGDARF
MAB_3325c|M.abscessus_ATCC_199 MRILIVGAGGVGSAAAFIAARRDFFEALVIADYDIARAQAVVDRLGDSRF
****:********* . *****.**: :*:.*** :** ..: :.*:**
MMAR_3555|M.marinum_M CSAALDAGSADAVAAAVRRHQITHVVNAVDPRFVMPIFEGALAAGADYLD
MUL_2788|M.ulcerans_Agy99 CSAALDAGCADAVAAAVRRHQITHVVNAVDPRFVMPIFEGALAAGADYLD
MSMEG_6311|M.smegmatis_MC2_155 TSAQVDATSADAVADLVREHQITHVMNAVDPRFVMPIFNGALAGGADYLD
MAB_3325c|M.abscessus_ATCC_199 MAARIDASCADDVTALCREHRITHALNAVDPRFVMSVFDGCFAAGVTYLD
:* :** .** *: *.*:***.:*********.:*:*.:*.*. ***
MMAR_3555|M.marinum_M MAMSLSQRHPEQPYQLTGVKLGDEQFAADDQWRAADRLALVGMGVEPGLS
MUL_2788|M.ulcerans_Agy99 MAISLSERHPEQPYQLTGVKLGDEQFAADDQWRAADRLALVGMGVEPGLS
MSMEG_6311|M.smegmatis_MC2_155 MAMSLSRRHPDRPYELTGVKLGDEQFGAAEDWEAAGRLALVGIGVEPGLS
MAB_3325c|M.abscessus_ATCC_199 MAMSLSHRHPDKPYELPGVMLGDEQFAATEKWKAAGLLALVGIGVEPGLS
**:***.***::**:*.** ******.* :.*.**. *****:*******
MMAR_3555|M.marinum_M DVFARYAADHLFTDIDELGTRDGANLTVDGYDFAPSFSIWTTIEECLNPP
MUL_2788|M.ulcerans_Agy99 DVFARYAADHLFTDIDELGTRDGANLTVDGYDFAPSFSIWTTIEECLNPP
MSMEG_6311|M.smegmatis_MC2_155 DVFARYAADHLFSEIDELGTRDGSSLVVEGCDFAPSFSIWTTIEECLNPP
MAB_3325c|M.abscessus_ATCC_199 DVFARYAADHLFSEIDELGTRDGSNLEVRGYRFAPSFSIWTTIEECLNPP
************::*********:.* * * ******************
MMAR_3555|M.marinum_M VIWEHDRGWFVTEPFSEPEVFDFPDGIGPVECVNVEHEEVLLMPRWVKAK
MUL_2788|M.ulcerans_Agy99 VIWEHDRGWFVTEPFSEPEVFDFPDGIGPVECVNVEHEEVLLMPRWVKAK
MSMEG_6311|M.smegmatis_MC2_155 VVWEADRGWFVTEPFSEPEVFDFPEGIGPVECVNVEHEEVLLMPRWVKCK
MAB_3325c|M.abscessus_ATCC_199 LIWERGKGFFTTEPFSEPEVFDFPGGIGPVECVNVEHEEVVLMPRWVNAR
::** .:*:*.************* ***************:******:.:
MMAR_3555|M.marinum_M RATFKYGLGTEFIEVLKTLRKLGLDRTEKITVGG----VQVSPRDVVAAC
MUL_2788|M.ulcerans_Agy99 RATFKYGLGTEFIEVLKTLRKLGLDRTEKITVGG----VQVSPRDVVAAC
MSMEG_6311|M.smegmatis_MC2_155 RATFKYGLGSEFIDVLKTLHKLGLDRTDKVRVPGANGPVEVSPRDVVAAC
MAB_3325c|M.abscessus_ATCC_199 RVTFKYGLGAEFINVLRTLHLVGLDSTAPVSVHANGGTAAVSPRDVVAAC
*.*******:***:**:**: :*** * : * . . **********
MMAR_3555|M.marinum_M LPDPATLGPKMHGKTCAGLWVTGTGKDGNPRSTYLYHVVDNQWSMNEYGH
MUL_2788|M.ulcerans_Agy99 LPDPATLGPKMHGKTCAGLWVTGTGKDGNPRSTYLYHVVDNQWSMNEYGH
MSMEG_6311|M.smegmatis_MC2_155 LPNPATLGPQMHGKTCAGLWVTGKGKDGAPRSTYLYHVVDNQWSMAEYGH
MAB_3325c|M.abscessus_ATCC_199 LPDPAGLGHLMRGATCAGLWVTGTGKDGRPREVYLHHVVDNEWTMARDGA
**:** ** *:* *********.**** **..**:*****:*:* . *
MMAR_3555|M.marinum_M QCVVWQTAINPVVALELLASGTWSGRGVLGPEAFDAVPFLELLSAYGAPW
MUL_2788|M.ulcerans_Agy99 QCVVWQTAINPVVALELLASGTWSGRGVLGPEAFDAVPFLELLSAYGAPW
MSMEG_6311|M.smegmatis_MC2_155 QCVVWQTAINPVVALELLASGMWSGAGVLGPEAFDAVPFLNLLKDYGSPW
MAB_3325c|M.abscessus_ATCC_199 QCVVWQTAVNPVVALELLAEGVWSGAGVLGPEAFDSLPFLDRLNTFGAPW
********:**********.* *** *********::***: *. :*:**
MMAR_3555|M.marinum_M GLQEVPARS
MUL_2788|M.ulcerans_Agy99 GLQEVPARS
MSMEG_6311|M.smegmatis_MC2_155 GIKEL----
MAB_3325c|M.abscessus_ATCC_199 GIQERAVAA
*::*