For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GQVSAVSTVLINAEPAAVLAAISDYQTVRPKILSSHYSGYQVLEGGQGAGTVATWKLQATKSRVRDVKAT VDVAGHTVIEKDANSSLVSNWTVAPAGTGSSVNLKTTWTGAGGVKGFFEKTFAPLGLRRIQDEVLENLKK HVEG*
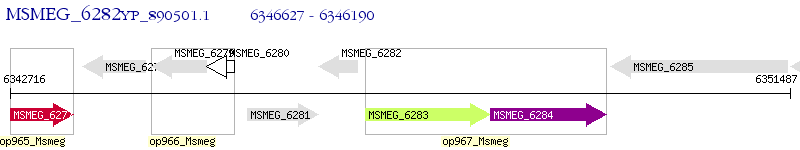
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6282 | - | - | 100% (145) | KanY protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3745c | - | 2e-63 | 78.62% (145) | hypothetical protein Mb3745c |
| M. gilvum PYR-GCK | Mflv_1285 | - | 6e-63 | 77.93% (145) | hypothetical protein Mflv_1285 |
| M. tuberculosis H37Rv | Rv3718c | - | 2e-63 | 78.62% (145) | hypothetical protein Rv3718c |
| M. leprae Br4923 | MLBr_02332 | - | 7e-60 | 73.61% (144) | hypothetical protein MLBr_02332 |
| M. abscessus ATCC 19977 | MAB_0317 | - | 1e-60 | 77.08% (144) | hypothetical protein MAB_0317 |
| M. marinum M | MMAR_5234 | - | 6e-62 | 78.47% (144) | hypothetical protein MMAR_5234 |
| M. avium 104 | MAV_0386 | - | 1e-64 | 81.25% (144) | KanY protein |
| M. thermoresistible (build 8) | TH_2290 | - | 2e-64 | 82.07% (145) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4309 | - | 4e-62 | 78.47% (144) | hypothetical protein MUL_4309 |
| M. vanbaalenii PYR-1 | Mvan_5530 | - | 2e-63 | 80.00% (145) | hypothetical protein Mvan_5530 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6282|M.smegmatis_MC2_155 -------MGQVSAVSTVLINAEPAAVLAAISDYQTVRPKILSSHYSGYQV
TH_2290|M.thermoresistible__bu -------MGQVSASNTVLIDAEPQAVLTAISDYETVRPKILTSHYSGYQV
Mflv_1285|M.gilvum_PYR-GCK -------MGQVSASSTVLIDSDPATVLDAVADYEAMRPKILSEHYSGYRV
Mvan_5530|M.vanbaalenii_PYR-1 -------MGQVSATSTVLIEADPATVLAAVADYQAMRPKILSAHYSGYRV
MMAR_5234|M.marinum_M -------MGQVSAASTILINAEPAAALAGVADYQNVRPKILSPQYSEYQV
MUL_4309|M.ulcerans_Agy99 MTEELRLMGQVSAASTILINAEPAAALAGVADYQNVRPKILSPQYSEYQV
MAV_0386|M.avium_104 -------MGQVSADSTILINADPAATLAAVADYQTVRPKILSPQYSDYQV
Mb3745c|M.bovis_AF2122/97 -------MGQVSAASTILINAEPTATLDALADYETVRPKILSPHYSEYQV
Rv3718c|M.tuberculosis_H37Rv -------MGQVSAASTILINAEPTATLDALADYETVRPKILSPHYSEYQV
MLBr_02332|M.leprae_Br4923 -------MGPVSAVSTILVNVEPVATLAAVADYQKMRPKILSPQYNEYQV
MAB_0317|M.abscessus_ATCC_1997 -------MGQVSASSSVLIDAAPEAVLAAVADYQNVRPQILSSHYRDYQV
** *** .::*:: * :.* .::**: :**:**: :* *:*
MSMEG_6282|M.smegmatis_MC2_155 LEGGQGAGTVATWKLQATKSRVRDVKATVDVAGHTVIEKDANSSLVSNWT
TH_2290|M.thermoresistible__bu LEGGQGAGTVVTWKLQATESRVRDVRATVDVAGRTVIEKDANSSMVTNWT
Mflv_1285|M.gilvum_PYR-GCK LEGGQGAGTVAEWKLQATKSRSRDVKANVDIAGRTVIEKDANSSMVTNWT
Mvan_5530|M.vanbaalenii_PYR-1 LEGGQGAGTVAEWKLQATKSRSRNVKADVDVAGRTVIEKDANSSMVTNWT
MMAR_5234|M.marinum_M LEGGQGQGTVAKWRLQATKSRVRDVQVGVDVAGHTVIEKDANSSMIINWT
MUL_4309|M.ulcerans_Agy99 LEGGQGQGTVAKWRLQATKSRVRDVQVGVDVAGHTVIEKDANSSMIINWT
MAV_0386|M.avium_104 LQGGQGAGTVAKWKLQATKSRVRDVQVDVDVAGHTVIEKDANSSMVTNWT
Mb3745c|M.bovis_AF2122/97 LEGGKGRGTVAKWRLQATQSRVRDVQVNVDVAGHTVIEKDMNSSMVTNWT
Rv3718c|M.tuberculosis_H37Rv LEGGKGRGTVAKWRLQATQSRVRDVQVNVDVAGHTVIEKDMNSSMVTNWT
MLBr_02332|M.leprae_Br4923 VQGGQGPGTVVKWKLQVTRSRVRDVQVNVDVAGHTVIEKDANSSMVTSWT
MAB_0317|M.abscessus_ATCC_1997 LEGGQGEGTVASWKLQATESRVRDIKSAVSVAGHTVIEKDANSTLVTTWT
::**:* ***. *:**.*.** *::: *.:**:****** **::: .**
MSMEG_6282|M.smegmatis_MC2_155 VAPAG-TGSSVNLKTTWTGAGGVKGFFEKTFAPLGLRRIQDEVLENLKKH
TH_2290|M.thermoresistible__bu VAPAGPAGSTVTVKTSWTGAGGVKGIFEKIFAPLGLRKIQAEVLENLKRE
Mflv_1285|M.gilvum_PYR-GCK VAPAG-PGSSVTVKTSWQGAGGIGGFFEKTFAPLGLRKIQAQVLENLKRE
Mvan_5530|M.vanbaalenii_PYR-1 VAPAG-PGSSVTVKTSWQGAGGIGGFFEKTFAPLGLRKIQAEVLGNLKRE
MMAR_5234|M.marinum_M VAPAG-PGCSVTVKTTWNGAGGVKGFFEKTFAPLGLKKIQGEVLANLKSE
MUL_4309|M.ulcerans_Agy99 VAPAG-PGCSVTVKTTWNGAGGVKGFFEKTFAPLGLKKIQGEVLANLKSE
MAV_0386|M.avium_104 VAPAG-PGSSVTVKTTWTGAGGVKGFFEKTFAPLGLKKIQGEVLANLKRE
Mb3745c|M.bovis_AF2122/97 VAPAG-PGSSVTVKTTWTGAGGVKGFFEKTFAPLGLKKIQAEVLSNLKTE
Rv3718c|M.tuberculosis_H37Rv VAPAG-PGSSVTVKTTWTGAGGVKGFFEKTFAPLGLKKIQAEVLSNLKTE
MLBr_02332|M.leprae_Br4923 VAPAG-PGSSVTMKTAWTGAGGVKGFFEKTFAPLGLKKIQAEVLANLKNE
MAB_0317|M.abscessus_ATCC_1997 VAPAG-PGSSVTTKTAWTGGGGIKGFFEKTFAPLGLKKIQAEVLANLKQV
***** .*.:*. **:* *.**: *:*** ******::** :** ***
MSMEG_6282|M.smegmatis_MC2_155 VEG---------
TH_2290|M.thermoresistible__bu IEQ----KGT--
Mflv_1285|M.gilvum_PYR-GCK VEG-----GKPA
Mvan_5530|M.vanbaalenii_PYR-1 VEGTIGIEGTQG
MMAR_5234|M.marinum_M LES---------
MUL_4309|M.ulcerans_Agy99 LES---------
MAV_0386|M.avium_104 LES---------
Mb3745c|M.bovis_AF2122/97 LEGDA-------
Rv3718c|M.tuberculosis_H37Rv LEGDA-------
MLBr_02332|M.leprae_Br4923 LER---------
MAB_0317|M.abscessus_ATCC_1997 VEKG--------
:*