For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALVVQKYGGSSVADAERIRRVAERIVETKKAGNDVVVVVSAMGDTTDDLLDLARQVSPAPPPREMDMLLT AGERISNALVAMAIESLGAQARSFTGSQAGVITTGTHGNAKIIDVTPGRLRDALDEGQIVLVAGFQGVSQ DSKDVTTLGRGGSDTTAVAVAAALDADVCEIYTDVDGIFTADPRIVPNARHLDTVSFEEMLEMAACGAKV LMLRCVEYARRYNVPIHVRSSYSDKPGTIVKGSIEDIPMEDAILTGVAHDRSEAKVTVVGLPDVPGYAAK VFRAVAEADVNIDMVLQNISKIEDGKTDITFTCARDNGPRAVEKLSALKSEIGFSQVLYDDHIGKVSLIG AGMRSHPGVTATFCEALAEAGINIDLISTSEIRISVLIKDTELDKAVSALHEAFGLGGDDEAVVYAGTGR *
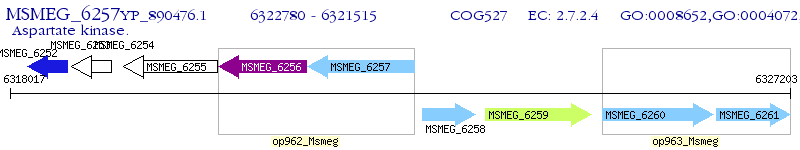
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6257 | - | - | 100% (421) | aspartate kinase |
| M. smegmatis MC2 155 | MSMEG_3774 | argB | 6e-09 | 27.83% (230) | acetylglutamate kinase |
| M. smegmatis MC2 155 | MSMEG_4621 | proB | 9e-05 | 24.00% (175) | gamma-glutamyl kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3736c | ask | 0.0 | 88.36% (421) | aspartate kinase |
| M. gilvum PYR-GCK | Mflv_1304 | - | 0.0 | 90.26% (421) | aspartate kinase |
| M. tuberculosis H37Rv | Rv3709c | ask | 0.0 | 88.36% (421) | aspartate kinase |
| M. leprae Br4923 | MLBr_02323 | ask | 0.0 | 85.51% (421) | aspartate kinase |
| M. abscessus ATCC 19977 | MAB_0343 | - | 0.0 | 87.17% (421) | aspartate kinase |
| M. marinum M | MMAR_5222 | ask | 0.0 | 86.94% (421) | aspartate kinase Ask |
| M. avium 104 | MAV_0393 | - | 0.0 | 87.41% (421) | aspartate kinase |
| M. thermoresistible (build 8) | TH_2282 | ask | 0.0 | 88.60% (421) | ASPARTOKINASE ASK (ASPARTATE KINASE) |
| M. ulcerans Agy99 | MUL_4297 | ask | 0.0 | 86.94% (421) | aspartate kinase |
| M. vanbaalenii PYR-1 | Mvan_5504 | - | 0.0 | 91.69% (421) | aspartate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1304|M.gilvum_PYR-GCK MRAALILYGISPRIAKGSPVALVVQKYGGSSVSDADRIRRVAERIVETKK
Mvan_5504|M.vanbaalenii_PYR-1 -------------------MALVVQKYGGSSVSDAERIRRVAERIVETKK
MSMEG_6257|M.smegmatis_MC2_155 -------------------MALVVQKYGGSSVADAERIRRVAERIVETKK
TH_2282|M.thermoresistible__bu -------------------VALVVQKYGGSSVSDAERIRRVAERIVETKK
MMAR_5222|M.marinum_M -------------------MALVVQKYGGSSVGNAERIRRVAERIVETKK
MUL_4297|M.ulcerans_Agy99 -------------------MALVVQKYGGSSVGNAERIRRVAERIVETKK
MAV_0393|M.avium_104 -------------------MALVVQKYGGSSVANADRIRRVAERIVATKK
Mb3736c|M.bovis_AF2122/97 -------------------MALVVQKYGGSSVADAERIRRVAERIVATKK
Rv3709c|M.tuberculosis_H37Rv -------------------MALVVQKYGGSSVADAERIRRVAERIVATKK
MLBr_02323|M.leprae_Br4923 -------------------MALVVQKYGGSSVADADRIRRVAERIVQTKK
MAB_0343|M.abscessus_ATCC_1997 -------------------MALVVQKYGGSSVATAERIRRVAERIVETKK
:************. *:********** ***
Mflv_1304|M.gilvum_PYR-GCK AGNDVVVVVSAMGDTTDELLDLAKQVSPAPPVREMDMLLTAGERISNALV
Mvan_5504|M.vanbaalenii_PYR-1 AGNDVVVVVSAMGDTTDELLDLAKQVSPAPPARELDMLLTAGERISNALV
MSMEG_6257|M.smegmatis_MC2_155 AGNDVVVVVSAMGDTTDDLLDLARQVSPAPPPREMDMLLTAGERISNALV
TH_2282|M.thermoresistible__bu AGNDVVVVVSAMGDTTDELLDLAKQVAPAPPAREMDMLLTAGERISNALV
MMAR_5222|M.marinum_M QGNDVVVVVSAMGDTTDDLMDLALQVCPVPPPRELDMLLTAGERISNALV
MUL_4297|M.ulcerans_Agy99 QGNDVVVVVSAMGDTTDDLMDLALQVCPVPPPRELDMLLTAGERISNALV
MAV_0393|M.avium_104 QGNDVVVVVSAMGDTTDDLMDLAQQVCPAPPPRELDMLLTAGERISNALV
Mb3736c|M.bovis_AF2122/97 QGNDVVVVVSAMGDTTDDLLDLAQQVCPAPPPRELDMLLTAGERISNALV
Rv3709c|M.tuberculosis_H37Rv QGNDVVVVVSAMGDTTDDLLDLAQQVCPAPPPRELDMLLTAGERISNALV
MLBr_02323|M.leprae_Br4923 QGNDIVVVVSAMGDTTDDLLDLAQQVCPEPPARELDMLLTAGERISNALV
MAB_0343|M.abscessus_ATCC_1997 NGNDVVVVVSAMGDTTDELLDLAQQVCPAPPPRELDMLLTAGERMSNALV
***:************:*:*** **.* ** **:*********:*****
Mflv_1304|M.gilvum_PYR-GCK AMAIESLGAQARSFTGSQAGVVTTGTHGNAKIIDVTPTRLRSALDEGQIV
Mvan_5504|M.vanbaalenii_PYR-1 AMAIESLGAEARSFTGSQAGVVTTGTHGNAKIIDVTPTRLRAALDEGQIV
MSMEG_6257|M.smegmatis_MC2_155 AMAIESLGAQARSFTGSQAGVITTGTHGNAKIIDVTPGRLRDALDEGQIV
TH_2282|M.thermoresistible__bu AMAIDSLGAQARSFTGSQAGVITTGSHGNAKIIDVTPGRVRSALDEGQIV
MMAR_5222|M.marinum_M AMAIESLGAHARSFTGSQAGVITTGTHGNAKIIEVTPGRLQAALDEGRIV
MUL_4297|M.ulcerans_Agy99 AMAIESLGAHARSFTGSQAGVITTGTHGNAKIIEVTPGRLQAALDEGRIV
MAV_0393|M.avium_104 AMAIESLGAQARSFTGSQAGVITTGTHGNAKIIDVTPGRLQAALDEGRIV
Mb3736c|M.bovis_AF2122/97 AMAIESLGAHARSFTGSQAGVITTGTHGNAKIIDVTPGRLQTALEEGRVV
Rv3709c|M.tuberculosis_H37Rv AMAIESLGAHARSFTGSQAGVITTGTHGNAKIIDVTPGRLQTALEEGRVV
MLBr_02323|M.leprae_Br4923 AMAIESFGAQARSFTGSQAGVITTGTHGNAKIIDVTPTRLQFALDEGRIV
MAB_0343|M.abscessus_ATCC_1997 AMAIHSLGASARSFTGSQAGVITTGKHGSAKIIDVTPGRLRTALDEGQVV
****.*:** ***********:***.**.****:*** *:: **:**::*
Mflv_1304|M.gilvum_PYR-GCK LVAGFQGVSQDTKDVTTLGRGGSDTTAVAVAAALNADVCEIYTDVDGVFT
Mvan_5504|M.vanbaalenii_PYR-1 LVAGFQGVSQDTKDVTTLGRGGSDTTAVAVAAALKADVCEIYTDVDGIFT
MSMEG_6257|M.smegmatis_MC2_155 LVAGFQGVSQDSKDVTTLGRGGSDTTAVAVAAALDADVCEIYTDVDGIFT
TH_2282|M.thermoresistible__bu LVAGFQGVSQDSKDVTTLGRGGSDTTAVALAAALKADVCEIYTDVDGIFT
MMAR_5222|M.marinum_M LVAGFQGVSQDSKDVTTLGRGGSDTTAVALAAALGADVCEIYTDVDGIFS
MUL_4297|M.ulcerans_Agy99 LVAGFQGVSQDSKDVTTLGRGGSDTTAVALAAALGADVCEIYTDVDGIFS
MAV_0393|M.avium_104 LVAGFQGVSQDTRDVTTLGRGGSDTTAVALAAALGADVCEIYTDVDGIFS
Mb3736c|M.bovis_AF2122/97 LVAGFQGVSQDTKDVTTLGRGGSDTTAVAMAAALGADVCEIYTDVDGIFS
Rv3709c|M.tuberculosis_H37Rv LVAGFQGVSQDTKDVTTLGRGGSDTTAVAMAAALGADVCEIYTDVDGIFS
MLBr_02323|M.leprae_Br4923 LVAGFQGVSQDTRDITTLGRGGSDTTAIAMAAALGADVCEIYTDVDGIFS
MAB_0343|M.abscessus_ATCC_1997 LVAGFQGVSQDSKDVTTLGRGGSDTTAVALAAALNADVCEIYTDVDGVFT
***********::*:************:*:**** ************:*:
Mflv_1304|M.gilvum_PYR-GCK ADPRIVPNARKLETVSFEEMLEMAAAGAKVLMLRCVEYARRFNLPIHVRS
Mvan_5504|M.vanbaalenii_PYR-1 ADPRIVPNARRLDTVSFEEMLEMAAAGAKVLMLRCVEYARRFDLPIHVRS
MSMEG_6257|M.smegmatis_MC2_155 ADPRIVPNARHLDTVSFEEMLEMAACGAKVLMLRCVEYARRYNVPIHVRS
TH_2282|M.thermoresistible__bu ADPRIVPNARHLDNVTFEEMLEMAAAGAKVLMLRCVEYARRYNVPIHVRS
MMAR_5222|M.marinum_M ADPRIVHNARKLDTVTFEEMLEMAACGAKVLMLRCVEYARRHNIPVHVRS
MUL_4297|M.ulcerans_Agy99 ADPRIVHNARKLDTVTFEEMLEMAACGAKVLMLRCVEYARRHNIPVHVRS
MAV_0393|M.avium_104 ADPRIVHNARKLDTVTFEEMLEMAACGAKVLMLRCVEYARRYNIPVHVRS
Mb3736c|M.bovis_AF2122/97 ADPRIVRNARKLDTVTFEEMLEMAACGAKVLMLRCVEYARRHNIPVHVRS
Rv3709c|M.tuberculosis_H37Rv ADPRIVRNARKLDTVTFEEMLEMAACGAKVLMLRCVEYARRHNIPVHVRS
MLBr_02323|M.leprae_Br4923 ADPRVVHNARKLDTVTFEEMLEMAACGAKVLMLRCVEYARRHNIPVHVRS
MAB_0343|M.abscessus_ATCC_1997 ADPRIVPNAARLDKVSFEEMLEMAAAGAKVLMLRCVEYARRYNVPVHVRS
****:* ** :*:.*:*********.***************.::*:****
Mflv_1304|M.gilvum_PYR-GCK SYTDRPGTLVTGSMEDIPMEDAILTGVAHDRGEAKVTVVGLPDVPGYAAQ
Mvan_5504|M.vanbaalenii_PYR-1 SYSDNPGTIVKGSIEDIPMEDAILTGVAHDRGEAKVTVVGLPDVPGYAAQ
MSMEG_6257|M.smegmatis_MC2_155 SYSDKPGTIVKGSIEDIPMEDAILTGVAHDRSEAKVTVVGLPDVPGYAAK
TH_2282|M.thermoresistible__bu SYSDKPGTLVTGSIEDIPMEDAILTGVAHDRSEARVTVVGIPDTPGFAAK
MMAR_5222|M.marinum_M SYSDKPGTLVVGSIKDIAMEDPILTGVAHDRSEAKVTIVGIPDIPGYAAK
MUL_4297|M.ulcerans_Agy99 SYSDKPGTLVVGSIKDIAMEDPILTGVAHDRSEAKVTIVGIPDIPGYAAK
MAV_0393|M.avium_104 SYSDKKGTVVVGSIKDKPMEDPILTGVAHDRSEAKVTVVGIPDIPGYAAR
Mb3736c|M.bovis_AF2122/97 SYSDRPGTVVVGSIKDVPMEDPILTGVAHDRSEAKVTIVGLPDIPGYAAK
Rv3709c|M.tuberculosis_H37Rv SYSDRPGTVVVGSIKDVPMEDPILTGVAHDRSEAKVTIVGLPDIPGYAAK
MLBr_02323|M.leprae_Br4923 SYADKPGTVIVGSIKDVPMEDPILTGVAHDRSEAKVTIVGIPDIPGYAAK
MAB_0343|M.abscessus_ATCC_1997 SYTDKPGTIVSGSMEDIPVEEAILTGVAHDRGEAKVTVVGIPDVPGYAAK
**:*. **:: **::* .:*:.*********.**:**:**:** **:**:
Mflv_1304|M.gilvum_PYR-GCK VFRACADADVNLDMVLQNISKVEDGKTDITFTCSRESGPGAVEKLTSLQN
Mvan_5504|M.vanbaalenii_PYR-1 VFRAVADADVNIDMVLQNISKVEDGKTDITFTCSRESGPGAVEKLTALQD
MSMEG_6257|M.smegmatis_MC2_155 VFRAVAEADVNIDMVLQNISKIEDGKTDITFTCARDNGPRAVEKLSALKS
TH_2282|M.thermoresistible__bu VFRAVADADINIDMVLQNASKVEDGKTDITFTCPRDAGPAAVEKLSSVQD
MMAR_5222|M.marinum_M VFRAVAEADVNIDMVLQNVSKVEDGRTDITFTCSRDSGPTAVAKLDLLKD
MUL_4297|M.ulcerans_Agy99 VFRAVAEADVNIDMVLQNVSKVEDGRTDITFTCSRDSGPTAVAKLDSLKD
MAV_0393|M.avium_104 VFRAVADADVNIDMVLQNVSKVEDGKTDITFTCSREAGPTAVAKLDALKD
Mb3736c|M.bovis_AF2122/97 VFRAVADADVNIDMVLQNVSKVEDGKTDITFTCSRDVGPAAVEKLDSLRN
Rv3709c|M.tuberculosis_H37Rv VFRAVADADVNIDMVLQNVSKVEDGKTDITFTCSRDVGPAAVEKLDSLRN
MLBr_02323|M.leprae_Br4923 VFRAVADADVNIDMVLQNVSKVEDGKTDITFTCSRDSGPIAVAKLGSLRD
MAB_0343|M.abscessus_ATCC_1997 VFRAVADAEINIDMVLQNVSKVEDGKTDITFTCPKENGPTAVEKLDSLKN
**** *:*::*:****** **:***:*******.:: ** ** ** ::.
Mflv_1304|M.gilvum_PYR-GCK EIGFTRVLYDDHIGKVSLIGAGMRSHPGVTATFCEALANAGINIDLISTS
Mvan_5504|M.vanbaalenii_PYR-1 EIGFTRVLYDDHIGKVSLIGAGMRSHPGVTATFCEALADAGINIDLISTS
MSMEG_6257|M.smegmatis_MC2_155 EIGFSQVLYDDHIGKVSLIGAGMRSHPGVTATFCEALAEAGINIDLISTS
TH_2282|M.thermoresistible__bu EIGFTKVLYDDHIGKVSLVGAGMRSHPGVTAKFCETLAAVGVNIDLISTS
MMAR_5222|M.marinum_M EVGFTQLLYDDHIGKLSLIGAGMRSHPGVTATFCEALAAVGVNIELISTS
MUL_4297|M.ulcerans_Agy99 EVGFTQLLYDDHIGKLSLIGAGMRSHPGVTATFCEALAAVGVNIELISTS
MAV_0393|M.avium_104 EIGFTQLLYDDHIGKVSLIGAGMRSHPGVTATFCEALAQVGVNIELISTS
Mb3736c|M.bovis_AF2122/97 EIGFSQLLYDDHIGKVSLIGAGMRSHPGVTATFCEALAAVGVNIELISTS
Rv3709c|M.tuberculosis_H37Rv EIGFSQLLYDDHIGKVSLIGAGMRSHPGVTATFCEALAAVGVNIELISTS
MLBr_02323|M.leprae_Br4923 EIGFTQLLYDDHIGKVSLIGAGMRSHPGVTATFCEALAAVGVNIELISTS
MAB_0343|M.abscessus_ATCC_1997 EIGFSQVLYDDHIGKVSLVGAGMRSHPGVTATFCEALAEAGVNIELISTS
*:**:::********:**:************.***:** .*:**:*****
Mflv_1304|M.gilvum_PYR-GCK EIRISVLIKDTELDRAVAALHEAFGLGGDEEAVVYAGTGR
Mvan_5504|M.vanbaalenii_PYR-1 EIRISVLIKDTELDRAVEALHDAFGLGGDEEAVVYAGTGR
MSMEG_6257|M.smegmatis_MC2_155 EIRISVLIKDTELDKAVSALHEAFGLGGDDEAVVYAGTGR
TH_2282|M.thermoresistible__bu EIRISVLIKDTDVDTAVAALHEAFELGGEETATVYAGTGR
MMAR_5222|M.marinum_M EIRISVLCRDTELDKAVVALHEAFGLGGEETATVYAGTGR
MUL_4297|M.ulcerans_Agy99 EIRISVLCRDTELDKAVVALHEAFGLGGEETATVYAGTGR
MAV_0393|M.avium_104 EIRISVLCRDTELDKAVVALHEAFGLGGAETATVYAGTGR
Mb3736c|M.bovis_AF2122/97 EIRISVLCRDTELDKAVVALHEAFGLGGDEEATVYAGTGR
Rv3709c|M.tuberculosis_H37Rv EIRISVLCRDTELDKAVVALHEAFGLGGDEEATVYAGTGR
MLBr_02323|M.leprae_Br4923 EIRISVLCRDTELDKAVFALHEAFGLGGEDEVTVYAGTGR
MAB_0343|M.abscessus_ATCC_1997 EIRISVLVKDDELDAAVQAIHNAFDLGSEEEATVYAGTGR
******* :* ::* ** *:*:** **. : ..*******