For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVLTGRTGLVALVCVLPIAVSPWPATAFGALLVLLLAAVGADVALAGSPRRLHVQRGGDHTARLGQTVDT VVEIHNPGRRPVRGQVRDAWPPSACAQPRVHPLDAAAAEQVRLVTRLQPVRRGDQRSETVTVRSIGPLGL AGRQAVHRVPHQVRILPPFLSRKHLPSRLARLRELDGAIPVLIRGQGTEFDSLREYVVGDDVRSIDWRAT ARRADVVVRTWRPERDRRVVIVLDTGRTSAGRVGVDPTASDPGGWPRLDWSMDAALLLAALASRAGDHVD FLAFDRVTRAGVFNASRTELMSALVGAMAPLQPALVESDATALVSTITRRVRGRSLVVLLTDLNASALDE GLMAVLPQLSARHQILLAAVSDPRVDDLAAGRSDAAAVYDAAAAERSRNDRRAIAARLRRRGVDVVDAPP EDLAPALADHYLALKATGKL
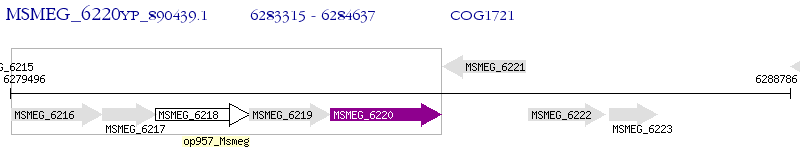
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6220 | - | - | 100% (440) | lipoprotein |
| M. smegmatis MC2 155 | MSMEG_6610 | - | 2e-14 | 27.16% (243) | protein of unknown function DUF58, putative |
| M. smegmatis MC2 155 | MSMEG_3148 | - | 4e-12 | 26.32% (285) | hypothetical protein MSMEG_3148 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3718 | - | 0.0 | 74.77% (440) | hypothetical protein Mb3718 |
| M. gilvum PYR-GCK | Mflv_1336 | - | 0.0 | 72.95% (440) | hypothetical protein Mflv_1336 |
| M. tuberculosis H37Rv | Rv3693 | - | 0.0 | 74.77% (440) | hypothetical protein Rv3693 |
| M. leprae Br4923 | MLBr_01809 | - | 3e-12 | 28.10% (242) | hypothetical protein MLBr_01809 |
| M. abscessus ATCC 19977 | MAB_0388c | - | 1e-163 | 65.11% (450) | hypothetical protein MAB_0388c |
| M. marinum M | MMAR_5203 | - | 0.0 | 75.45% (440) | hypothetical protein MMAR_5203 |
| M. avium 104 | MAV_0411 | - | 0.0 | 76.14% (440) | lipoprotein |
| M. thermoresistible (build 8) | TH_1716 | - | 0.0 | 76.14% (440) | POSSIBLE CONSERVED MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4276 | - | 0.0 | 75.45% (440) | hypothetical protein MUL_4276 |
| M. vanbaalenii PYR-1 | Mvan_5456 | - | 0.0 | 74.15% (441) | hypothetical protein Mvan_5456 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6220|M.smegmatis_MC2_155 MVLTGRTGLVALVCVLPIAVSPWPATAFGALLVLLLAAVGADVALAGSPR
TH_1716|M.thermoresistible__bu MIVTGRTALVAAVAILPIAVSPWPAMAFAALLSLLVVAVVVDAALAANPR
Mflv_1336|M.gilvum_PYR-GCK MILTGRAGLLALLCAVPVLSSPSPAVVFWTLAGLLTIAIVCDVALAGSPR
Mvan_5456|M.vanbaalenii_PYR-1 MVLTGRAGLIALLCALPVVLSPYPAATFAVLAALLAVAVLADVALAGSPR
Mb3718|M.bovis_AF2122/97 MILTGRTGLLALICVLPIALSPWPARAFVMLLVALAVAVTVDTLLAASTR
Rv3693|M.tuberculosis_H37Rv MILTGRTGLLALICVLPIALSPWPARAFVMLLVALAVAVTVDTLLAASTR
MMAR_5203|M.marinum_M MILTGRTGLMALIGVLPIALSPWPPRAFGMLLLALAVAVAIDVALAASPR
MUL_4276|M.ulcerans_Agy99 MILTGRTGLMALIGVLPIALSPWPPRAFGMLLLALAVAVAIDVALAASPR
MAV_0411|M.avium_104 MILTGRTGLVALICVLPIALSPWPAMVFTALLAALAVAVAVDAALAASPA
MAB_0388c|M.abscessus_ATCC_199 MVLTGRAGLVAALSTLLILVSPWPVRLFWLLLLALALLIAVDIAAAASPR
MLBr_01809|M.leprae_Br4923 ---------------------------------------MIDPNPAAKPG
* *...
MSMEG_6220|M.smegmatis_MC2_155 RLHVQRGGDHTARLGQTVDTVVEIHNPGRRPVRGQVRDAWPPSACAQPRV
TH_1716|M.thermoresistible__bu QLLFSRSGESTARLGQSVDALLEVANGGGRRLRGRLRDAWPPSARAEPRL
Mflv_1336|M.gilvum_PYR-GCK ALRLTRGGPTSARLGQPVTVDLTVENGGRARFRGVVRDAWAPSARAHPRT
Mvan_5456|M.vanbaalenii_PYR-1 ALELTRGGPTSARLGQPVTAELTVRNSGRARFRGLVRDAWAPSARGAPRV
Mb3718|M.bovis_AF2122/97 KLRFTRSPYTSARLGQPVDASLLLCNGGRRRFRGQVRDAWPPSARAQPHT
Rv3693|M.tuberculosis_H37Rv KLRFTRSPYTSARLGQPVDASLLLCNGGRRRFRGQVRDAWPPSARAQPHT
MMAR_5203|M.marinum_M KLSYTRGANSSIRLGQPADAALVIHNVGRRRFRGLVRDAWPPSARAQPRT
MUL_4276|M.ulcerans_Agy99 KLSYTRGANSSIRLGQPADAALVIHNVGRRRFRGLVRDAWPPSARAQPRT
MAV_0411|M.avium_104 RLRCIRSPDRSARLGQHADAALLIHNDGPRRFRGQIRDAWPPSARARPRL
MAB_0388c|M.abscessus_ATCC_199 RLTFTRSGPDSIRLGESAEIVLLAQNPG-RRLRGWVRDSWAPSMRAAPRA
MLBr_01809|M.leprae_Br4923 VFHPPSMQ------------------------RGQIDD---PKLSAALRT
: ** : * *. . :
MSMEG_6220|M.smegmatis_MC2_155 HPLDAAAAEQVRLVTRLQPVRRGDQRSETVTVRSIGPLGLAGRQAVHRVP
TH_1716|M.thermoresistible__bu HALHLDPGQRVRLRTTLRPTRRGDQHSALVTVRTLGPAGLAGRQRSHRVP
Mflv_1336|M.gilvum_PYR-GCK HAVDLPVGQRVRISTELTPARRGDQVSALVTARSVGPLGLAGRQGSHRVP
Mvan_5456|M.vanbaalenii_PYR-1 HRVNIAAGQRVLLETELRPVRRGDQVSALVTARAIGPLGLAGRQGSHRVP
Mb3718|M.bovis_AF2122/97 HDVDVAAGQRQQVHTALRPVRRGDQRAAMVTARSIGPLGLAGRQSSQSVP
Rv3693|M.tuberculosis_H37Rv HDVDVAAGQRQQVHTALRPVRRGDQRAAMVTARSIGPLGLAGRQSSQSVP
MMAR_5203|M.marinum_M HPINIAAGERQQVNTALQPVRRGDQHCAAVTARSIGPLGLAGRQRSQVVP
MUL_4276|M.ulcerans_Agy99 HPINIAAGERQQVNTALQPVRRGDQHCAAVTARSIGPLGLAGRQRSQVVP
MAV_0411|M.avium_104 HRVDIPAGQHQHIATRLEPVRRGDQRATVVTARSIGPLGLAGRQGSQSVP
MAB_0388c|M.abscessus_ATCC_199 HRVDLRSGERTQIVTTLRPLRRGDHKPATVTVRTIGPLGIAGRQGRHDVP
MLBr_01809|M.leprae_Br4923 LELTVKR--------KLDGVLHGDHLG-----------------------
: * :**:
MSMEG_6220|M.smegmatis_MC2_155 HQVRILPPFLSRKHLPSRLARLRELDGAIPVLIRGQGTEFDSLREYVVGD
TH_1716|M.thermoresistible__bu GRVRVLPPFLSRKHLPARLARLRDLEGAVPVLIRGQGTEFDSLREYVVGD
Mflv_1336|M.gilvum_PYR-GCK WQVRILPPFLSRKHLPSRLARLRELEGMTPVLIRGQGTEFDSLREYVVGD
Mvan_5456|M.vanbaalenii_PYR-1 WQIRILPPFLSRKHLPSRLARLRELEGMTPVLIRGQGTEFDSLREYVVGD
Mb3718|M.bovis_AF2122/97 GLVRVLPPFLSRKHLPSRLAKLREIDGLLPTLIRGQGTEFDSLREYVVGD
Rv3693|M.tuberculosis_H37Rv GLVRVLPPFLSRKHLPSRLAKLREIDGLLPTLIRGQGTEFDSLREYVVGD
MMAR_5203|M.marinum_M GQVRVLPPFLSRKHLPSRLAKLREIDGLLPTLIRGQGTEFDSLREYVVGD
MUL_4276|M.ulcerans_Agy99 GQVRVLPPFLSRKHLPSRLAKLREIDGLLPTLIRGQGTEFDSLREYVVGD
MAV_0411|M.avium_104 GQLRVLPPFLSRKHLPSRLARLREIDGLLPTLVRGQGTEFDSLREYVVGD
MAB_0388c|M.abscessus_ATCC_199 WQLRVLPPFLSRKHLPSRLAKLRELEGAIPVLIRGQGTEFDSLREYVVGD
MLBr_01809|M.leprae_Br4923 -------------------------------LISGPGSEPGESRVYQPGD
*: * *:* .. * * **
MSMEG_6220|M.smegmatis_MC2_155 DVRSIDWRATARRADVVVRTWRPERDRRVVIVLDTGRTSAGRVGVDPTAS
TH_1716|M.thermoresistible__bu DVRSIDWRATARKDDVVVRTWRPERDQRVVIALDTGRTSAGRVGVDPTGS
Mflv_1336|M.gilvum_PYR-GCK DVRSIDWRATARRADVVVRTWRPERDRRVLIVLDTGRTSAGRVGVDPTLL
Mvan_5456|M.vanbaalenii_PYR-1 DVRSIDWRATARRADVVVRTWRPERDRRMLIVLDTGRTSAGRVGVDPTGS
Mb3718|M.bovis_AF2122/97 DVRSIDWRASARRADVMVRTWRPERDRRVVIVLDTGRMAAGRVGVDPTAA
Rv3693|M.tuberculosis_H37Rv DVRSIDWRASARRADVMVRTWRPERDRRVVIVLDTGRMAAGRVGVDPTAA
MMAR_5203|M.marinum_M DVRSIDWRASARRADVMVRTWRPERDRRVVLVLDTGRMAAGRVGVDPTDA
MUL_4276|M.ulcerans_Agy99 DVRSIDWRASARRADVMVRTWRPERDRRVVLVLDTGRMAAGRVGVDPTDA
MAV_0411|M.avium_104 DVRSIDWRATARRADVVVRTWRPERDRRVVIVLDTGRTAAGRVGVDPTAA
MAB_0388c|M.abscessus_ATCC_199 DVRSIDWRASARRNDVVVRTWRPERDRRILVVLDTGRTSAGRIGVDPTSG
MLBr_01809|M.leprae_Br4923 DVRRMDWAVTARTTHPHVRQMIADRELETWMVIDMS----ASLDFGTTIC
*** :** .:** . ** .:*: . :.:* . . :....*
MSMEG_6220|M.smegmatis_MC2_155 DPG-GWPRLDWSMDAALLLAALASRAGDHVDFLAFD--RVTRAGVFNASR
TH_1716|M.thermoresistible__bu EPG-GWPRLDWSMDAALLLAALASRAGDRVDFLAHD--RELRAAVFNAPR
Mflv_1336|M.gilvum_PYR-GCK DPG-GWPRLDWSMDAALLLAALAARAGDHVDLLAHD--RETRAAVVNASR
Mvan_5456|M.vanbaalenii_PYR-1 DPATGWPRLDWSMDAALLLAALAARAGDHVDFLAHD--RETCAAVVNASR
Mb3718|M.bovis_AF2122/97 DPA-GWPRLDWSMDAALLLAALASRAGDHVDFLAHD--RISRAGVFGASR
Rv3693|M.tuberculosis_H37Rv DPA-GWPRLDWSMDAALLLAALASRAGDHVDFLAHD--RISRAGVFGASR
MMAR_5203|M.marinum_M DPA-GWPRLDWSMDAALLLAALASRAGDHVDFLAHD--RVSRAAVFGASR
MUL_4276|M.ulcerans_Agy99 DPA-GWPRLDWSMDAALLLAALASRAGDHVDFLAHD--RVSRAAVFGASR
MAV_0411|M.avium_104 DPA-GWPRLDWSMDAALLLAALASRAGDHVDFLAHD--RVGRAGVFGASR
MAB_0388c|M.abscessus_ATCC_199 DPS-GWPRLDWAMDAALLLVALASRAGDRVDFLAHD--RGVRAQVSGASR
MLBr_01809|M.leprae_Br4923 EKR------DLAVAAAAAITFLNSGGGNRLGALICNGARMTRVPARSGRQ
: * :: ** :. * : .*:::. * : * . . .. :
MSMEG_6220|M.smegmatis_MC2_155 TELMSALVGAMAPLQPALVESDATALVSTITRRVRGRSLVVLLTDLNASA
TH_1716|M.thermoresistible__bu TELLARLVDAMAPLQPALVESDASALVAEVQRRVRRRALVVLLTDLNASA
Mflv_1336|M.gilvum_PYR-GCK TELLAQLVSAMAPLEPALVESDARGMVAAVQRRVRRAALVVLLTDLNASA
Mvan_5456|M.vanbaalenii_PYR-1 TELLAQLVAAMAPLEPSLIESDARAMVAAVQRRVRRRALVVLLTDLNASA
Mb3718|M.bovis_AF2122/97 SELLAQLVDAMAPLRPALIESDWHAMIATILRRTRRRSLVVLLTDLNATA
Rv3693|M.tuberculosis_H37Rv SELLAQLVDAMAPLRPALIESDWHAMIATILRRTRRRSLVVLLTDLNATA
MMAR_5203|M.marinum_M TELLAQLVDAMAPLQPALVESDWRSMVATIARRTRRRSLVVLLTDLNATA
MUL_4276|M.ulcerans_Agy99 TELLAQLVDAMAPLQPALVESDWRSMVATIARRTRRRSLVVLLTDLNATA
MAV_0411|M.avium_104 TELLAQLVDAMAPLQPTLVETDWRAMVSAIALRARRRSLVVLLTDLNAAA
MAB_0388c|M.abscessus_ATCC_199 NELLPLVVNAMAPLEPSLVESDARGLVSTIRRRSRHRSLIVLLTDLNPAA
MLBr_01809|M.leprae_Br4923 HEQTLLRTIATTPKAPVGVRGDLTVAIDALRRPERRRGMAVIISDFLG--
* . * :* * :. * : : * .: *:::*:
MSMEG_6220|M.smegmatis_MC2_155 LDEGLMAVLPQLSARHQILLAAVSDPRVDDLAAGR----------SDAAA
TH_1716|M.thermoresistible__bu LDEGLMGVLPQLTARHKVLLAAVADPRVDQLAAGR----------ADAAQ
Mflv_1336|M.gilvum_PYR-GCK LDEGLLSVLPQLTAKHRVIVAAVADPRVEKLAAGR----------ADAVE
Mvan_5456|M.vanbaalenii_PYR-1 IDEGLLKVLPQLTAKHRVIVAAVADPRVDTLAAGR----------ADAVE
Mb3718|M.bovis_AF2122/97 LDEGLLPVLPQLSARHHVLVAAVADPRVDQLAAGR----------SDAAA
Rv3693|M.tuberculosis_H37Rv LDEGLLPVLPQLSARHHVLVAAVADPRVDQLAAGR----------SDAAA
MMAR_5203|M.marinum_M LDEGLLPVLPQLSAKHQVVVAAVADPRVDQLAIGR----------SDAAA
MUL_4276|M.ulcerans_Agy99 LDEGLLPVLPQLSAKHQVVVAAVADPRVDQLAIGR----------SDAAA
MAV_0411|M.avium_104 LDEGLLPVLPQLSAKHHLLVAAVADPRVDQMAAGR----------SDAAA
MAB_0388c|M.abscessus_ATCC_199 LEEGLLPLLSQLTTHHHLLVAAVSDPRVEDMVTPANATGPGNIRSLDAEQ
MLBr_01809|M.leprae_Br4923 -PINWMRPLRAIAARHEVLGIEVLDPRDVALPDIG--EVVLQDAETGVTR
. : * ::::*.:: * *** : ..
MSMEG_6220|M.smegmatis_MC2_155 VYDAAAAERSRNDRRA------IAARLRRRGVDVVDAPPEDLAPALADHY
TH_1716|M.thermoresistible__bu VYDAASAERSRNERRA------IAMRLRRHGVEVVDAPPEELAPALADHY
Mflv_1336|M.gilvum_PYR-GCK VYDAAAAERARNDRSA------VASRLRSLGVDVVDSPPEELAPDLADHY
Mvan_5456|M.vanbaalenii_PYR-1 VYDAAAAERARNDRRA------VASRLRAMGVDVVDAPPEDLAPDLADHY
Mb3718|M.bovis_AF2122/97 VYDAAAAERARNDRRA------IASQLRRGGVDVIDAPPAEIAPGLADRY
Rv3693|M.tuberculosis_H37Rv VYDAAAAERARNDRRA------IASQLRRGGVDVIDAPPAEIAPGLADRY
MMAR_5203|M.marinum_M VYDAAAAERARNDRGT------IAARLRRSGVEVVDAPPTEIAPALADRY
MUL_4276|M.ulcerans_Agy99 VYDAAAAERARNDRGT------IAARLRRSGVEVVDAPPTEIAPALADRY
MAV_0411|M.avium_104 VYDAAAAERSRNERAA------IAARLRHGGVEVVDAAPAELAPALADHY
MAB_0388c|M.abscessus_ATCC_199 VYDAAAAERTRGGRRR------IVTLLRRQGVEVVDAPPDEIAPALADRY
MLBr_01809|M.leprae_Br4923 EFTIDAALRDDFARAAAAHCADVSRSLRNCGAPLMSLRTDRDWIADIVRF
: :* * * : ** *. ::. . ::
MSMEG_6220|M.smegmatis_MC2_155 LALKATGKL----
TH_1716|M.thermoresistible__bu LAIKAAGRL----
Mflv_1336|M.gilvum_PYR-GCK LAMKATGRL----
Mvan_5456|M.vanbaalenii_PYR-1 LAMKAAGRL----
Mb3718|M.bovis_AF2122/97 LAMKATGRL----
Rv3693|M.tuberculosis_H37Rv LAMKATGRL----
MMAR_5203|M.marinum_M LAMKATGRL----
MUL_4276|M.ulcerans_Agy99 LAMKATGRL----
MAV_0411|M.avium_104 LAMKATGRL----
MAB_0388c|M.abscessus_ATCC_199 LSLKASGRL----
MLBr_01809|M.leprae_Br4923 VESRRRGALAGRQ
: : * *