For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTDGGAPAPFDPDDPARGAITRGSIARVGSATAITALCGYAVLYLAARDLEPAGFSVFAVFWGAFGLVTG AANGLLQEATREVRSVQVAPRVSGVSGPRTNPMRVAAAVGVVAALVIAGTSPLWSGHVFVESRALSVALL SVGLAGFCLHATLLGMLAGIDRWTEYGALMVTDAGIRVGVAAASFVLGWGLGGYLWATVAGAGAWLILLL TAPAARVAARQVTAGGTWTFVRGTAHSIAAAGASAILVMGFPVLLKATSADLGAAGGVVILAVTLTRAPL LVPLTAMQGNLIAHFVDQARLRALLMPAAVVAVIGAAGVVAAGLIGPWLLKVGFGDEYRADGTLLAWLTA AAVAIAMLTLTGAAAVAAALHRTYALGWIIATVAAVGLLLLPLELSTRTVVALLCGPLVGIAVHLVGLAR FSRVSG
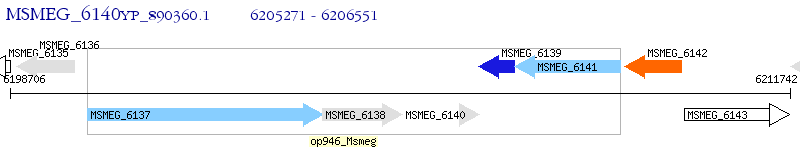
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6140 | - | - | 100% (426) | membrane protein |
| M. smegmatis MC2 155 | MSMEG_5946 | - | 2e-06 | 25.00% (196) | hypothetical protein MSMEG_5946 |
| M. smegmatis MC2 155 | MSMEG_4886 | - | 3e-05 | 24.71% (340) | major facilitator family protein transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3654 | - | 1e-164 | 70.88% (419) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_1402 | - | 1e-161 | 71.36% (405) | putative integral membrane protein |
| M. tuberculosis H37Rv | Rv3630 | - | 1e-164 | 70.64% (419) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0508c | - | 1e-137 | 63.37% (404) | hypothetical protein MAB_0508c |
| M. marinum M | MMAR_5130 | - | 1e-166 | 73.83% (405) | hypothetical protein MMAR_5130 |
| M. avium 104 | MAV_0526 | - | 1e-167 | 73.85% (413) | hypothetical protein MAV_0526 |
| M. thermoresistible (build 8) | TH_0619 | - | 1e-163 | 71.67% (406) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4207 | - | 1e-162 | 73.37% (398) | hypothetical protein MUL_4207 |
| M. vanbaalenii PYR-1 | Mvan_5388 | - | 1e-169 | 75.80% (405) | putative integral membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3654|M.bovis_AF2122/97 MAVGAAAVTEVGDTASPVGSSGASGGAIASGSVARVGTATAVTALCGYAV
Rv3630|M.tuberculosis_H37Rv MAVGAAAVTEVGDTASPVGSSGASGGAIASGSVARVGTAAAVTALCGYAV
MMAR_5130|M.marinum_M -------MTEVGGTT---GAAGASGGAIARGSVARVGTATAVTALCGYTV
MUL_4207|M.ulcerans_Agy99 --------------------------------MARVGTATAVTALCGYTV
MAV_0526|M.avium_104 -------MTEI----------SASQGPVARGSMARVGTATAVTALCGYAV
MSMEG_6140|M.smegmatis_MC2_155 -------MTDGGAPAP-FDPDDPARGAITRGSIARVGSATAITALCGYAV
Mflv_1402|M.gilvum_PYR-GCK -------MTDAT---------AGPSGAVSRASVVRVGVATALSALCGYAV
Mvan_5388|M.vanbaalenii_PYR-1 -------MTDAT---------AGPSGAVTRGSVVRVGAATAMSAMCGYAV
TH_0619|M.thermoresistible__bu -------VTDAT----------PATGAITRASVMRVGVATALSALCGYAV
MAB_0508c|M.abscessus_ATCC_199 -------MTDAG--------------PSTRSSVARIGAATAVSAICGYAV
: *:* *:*::*:***:*
Mb3654|M.bovis_AF2122/97 IYLAARNLAPNGFSVFGVFWGAFGLVTGAANGLLQETTREVRSLGYLD--
Rv3630|M.tuberculosis_H37Rv IYLAARNLAPNGFSVFGVFWGAFGLVTGAANGLLQETTREVRSLGYLD--
MMAR_5130|M.marinum_M IYLAARDLAPSGFSVFGVFWGAFGLVSGAANGLLQEATREVRSAQYLD--
MUL_4207|M.ulcerans_Agy99 IYLAARDLAPSGFSVFGVFWGAFGLVSGTANGLLQEATREVRSAQYLD--
MAV_0526|M.avium_104 IYLAARDLAPSGFSVFGVFWGAFGLVTGAANGLLQETTREVRVMPYLE--
MSMEG_6140|M.smegmatis_MC2_155 LYLAARDLEPAGFSVFAVFWGAFGLVTGAANGLLQEATREVRSVQVAPRV
Mflv_1402|M.gilvum_PYR-GCK LYLAARDLEPAGFSVFGVFWGAFGLVSGAAFGLLQETTREVRHPSAGT--
Mvan_5388|M.vanbaalenii_PYR-1 LYLAARDLEPAGFSVFGVFWGAFGLASGAAFGLLQETTREVRAPSWNP--
TH_0619|M.thermoresistible__bu MYLAARDLAPEGFSVFSVFWGAFGLVTGAANGLLQEATREVRSVRHTP--
MAB_0508c|M.abscessus_ATCC_199 LYLAARELGPALFSVFGVFWGAFGLVTGAANGLLQETTREVRLSLAQR--
:*****:* * ****.********.:*:* *****:*****
Mb3654|M.bovis_AF2122/97 VSADGRRTHPLRVSGMVGLGSLVVIAGSSPLWSGRVFAEARWLSVALLSI
Rv3630|M.tuberculosis_H37Rv VSADGRRTHPLRVSGMVGLGSLVVIAGSSPLWSGRVFAEARWLSVALLSI
MMAR_5130|M.marinum_M VVPEGGRTHPLRIAGMVGLASAVVIAGSSPLWSGRVFVEDRWLSVGLLSV
MUL_4207|M.ulcerans_Agy99 VVPEGGRTHPLRIAGMVGLASAVVIAGSSPLWSGRVFVEDRWLSVGLLSV
MAV_0526|M.avium_104 VAP-VKRTHPLRVAMLLGAAAAVVIAGSSPLWSGRVFVEARPLSVLLLSV
MSMEG_6140|M.smegmatis_MC2_155 SGVSGPRTNPMRVAAAVGVVAALVIAGTSPLWSGHVFVESRALSVALLSV
Mflv_1402|M.gilvum_PYR-GCK ---AHRHTRPLAVAAAVGVAAAAAIGVSAPLWSAHVFVDARMLSVALLCA
Mvan_5388|M.vanbaalenii_PYR-1 ---DGTYTRPMRVAAGVGVGAAAVIAVTSPLWSGHVFVDARGLSVALLSA
TH_0619|M.thermoresistible__bu -DPAVRRTRPMLIATLVAGLAAAAIVGTAPVWAGHVFAEARPLSVLLLAA
MAB_0508c|M.abscessus_ATCC_199 DRSPLATTLPVRIASAFAIASAVLIVGTAPLWASHVFTEFRWLSVALLAG
* *: :: .. : * ::*:*:.:**.: * *** **.
Mb3654|M.bovis_AF2122/97 GLAGFCLHATLLGMLAGTNRWTQYGALMVADAVIRVVVAAATFVIGWQLV
Rv3630|M.tuberculosis_H37Rv GLAGFCLHATLLGMLAGTNRWTQYGALMVADAVIRVVVAAATFVIGWQLV
MMAR_5130|M.marinum_M GLAGFCLHATLLGMLAGTNRWSHYGSLMVTDAIIRVAVSAATFAIGWGLV
MUL_4207|M.ulcerans_Agy99 GLAGFCLHATLLGMLAGTNRWSHYGSLMVTDAIIRVAVSAATFAIGWGLV
MAV_0526|M.avium_104 GLAGFCVHATLLGMLAGTNEWTRYGALMVTDAVIRVMVAAATVVLGWRLV
MSMEG_6140|M.smegmatis_MC2_155 GLAGFCLHATLLGMLAGIDRWTEYGALMVTDAGIRVGVAAASFVLGWGLG
Mflv_1402|M.gilvum_PYR-GCK GLAGFCVHTTLLGALAGMGRWSDYGALMVTDAALRVVVAAAGFAAGWGLG
Mvan_5388|M.vanbaalenii_PYR-1 GLAGFCLHTTLLGALAGMNRWSEYGALMVTDAVLRVVVAAATFALAWGLG
TH_0619|M.thermoresistible__bu GVAGFCLHATLLGMLAGVNRWTQYGSLMVTDAAIRVVVAAATFAVGWGLV
MAB_0508c|M.abscessus_ATCC_199 GLAGFCMHATTLGALAGSSRWTQYGVLMVTDAGIRLVVAIVTFAATLGLV
*:****:*:* ** *** ..*: ** ***:** :*: *: . .. *
Mb3654|M.bovis_AF2122/97 GFIWATVAGSVAWLIMLMTSPPTRAAARLMTPGATATFLRGAAHSIIAAG
Rv3630|M.tuberculosis_H37Rv GFIWATVAGSVAWLIMLMTSPPTRAAARLMTPGATATFLRGAAHSIIAAG
MMAR_5130|M.marinum_M GFLWATVAGSVSWLILLLASPVARATARLLTPGNTATFLRGAAHSITAAG
MUL_4207|M.ulcerans_Agy99 GFLWATVAGSVSWLILLLASPVARATARLLTPGNTATFLRGAAHSITAAG
MAV_0526|M.avium_104 GFLWATVAGAVAWLILLAASPATRATARLLTPGGTATFLRGAAHSITAAG
MSMEG_6140|M.smegmatis_MC2_155 GYLWATVAGAGAWLILLLTAPAARVAARQVTAGGTWTFVRGTAHSIAAAG
Mflv_1402|M.gilvum_PYR-GCK GYLWATVAGAFSWLLMLLVSPATRAAARVQTTGGTATFLRGAGHSMAAAG
Mvan_5388|M.vanbaalenii_PYR-1 GYLWATVAGSTAWLLMMLVSPATRAAARLLTPGGTATFLRGAGHSIAAAG
TH_0619|M.thermoresistible__bu GFLWATVAGAVAWLLMLLVSPSARQAARVRTAGGTVAFLRGASHSIAAAG
MAB_0508c|M.abscessus_ATCC_199 GYLWATVSGSMSWLLLTLVSKPTRDALKVPALVDTMEFLRGARHSIAAAA
*::****:*: :**:: .: :* : : : * *:**: **: **.
Mb3654|M.bovis_AF2122/97 ASAILVMGFPVLLKLTSNELGAQGGVVILAVTLTRAPLLVPLTAMQGNLI
Rv3630|M.tuberculosis_H37Rv ASAILVMGFPVLLKLTSNELGAQGGVVILAVTLTRAPLLVPLTAMQGNLI
MMAR_5130|M.marinum_M ASAILVMGFPVLLKLTSNELGPQGGVIILAVTLTRAPLLVPLTAMQGNLI
MUL_4207|M.ulcerans_Agy99 ASAILVMGFPVLLKLTSNELGPQGGVIILAVTLTRAPLLVPLTAMQGNLI
MAV_0526|M.avium_104 ASAILVMGFPVLLKLTSAELGAQGGVIILAVTLTRAPLLVPLTAMQGNLI
MSMEG_6140|M.smegmatis_MC2_155 ASAILVMGFPVLLKATSADLGAAGGVVILAVTLTRAPLLVPLTAMQGNLI
Mflv_1402|M.gilvum_PYR-GCK ASAVLVMGFPVLLAATSGDLGATGGVVILAVTLTRAPLMVPLTAMQGNLI
Mvan_5388|M.vanbaalenii_PYR-1 ASAVLVMGFPVLLKATSGDLGAAGGVVILAVTLTRAPLLVPLTAMQGNLI
TH_0619|M.thermoresistible__bu ASAVLVTGFPVLLKATYGDLGAAGGVVILAVMLTRAPLLIPLTAMQGNLI
MAB_0508c|M.abscessus_ATCC_199 ASAVLVMGFPVLLQSTAGDLGTTGGVVILAVTLTRAPLLVPLTALQGNLI
***:** ****** * :**. ***:**** ******::****:*****
Mb3654|M.bovis_AF2122/97 AHFVDERTERIRALIAPAALIGGVGAVGMLAAGVVGPWIMRVAFGSEYQS
Rv3630|M.tuberculosis_H37Rv AHFVDERTERIRALIAPAALIGGVGAVGMLAAGVVGPWIMRVAFGSEYQS
MMAR_5130|M.marinum_M AHFVDESAQRLRALISPAALIGGIGVIGVLAAGAVGPWLLRSAFGQQYQA
MUL_4207|M.ulcerans_Agy99 AHFVDESAQRLRALISPAALIGGIGVIGVLAAGAVGPWLLRSAFGQQYQA
MAV_0526|M.avium_104 AHFVDERSDRVRALIGPAAIVGAIGAVGVLAAGVLGPWVLRVAFGPQYQA
MSMEG_6140|M.smegmatis_MC2_155 AHFVDQ--ARLRALLMPAAVVAVIGAAGVVAAGLIGPWLLKVGFGDEYRA
Mflv_1402|M.gilvum_PYR-GCK AYFIDQRAHRLKALLAPSALVAALGAGGVVLAAVFGPWLLREAFGAQYRA
Mvan_5388|M.vanbaalenii_PYR-1 AHFVDQREHRLKALVAPAALVGALGAAGVALAAAFGPWLLREAFGAQYHA
TH_0619|M.thermoresistible__bu AHFVDERARRLRALVAPVGVVVTVGAVGVVGAGLVGPWLLRVAFGPDYVA
MAB_0508c|M.abscessus_ATCC_199 AHFVDEQDRRLRALATPAAIILGVGAAGIALAASIGPWLLRTVFDPAYQA
*:*:*: *::** * .:: :*. *: *. .***::: *. * :
Mb3654|M.bovis_AF2122/97 SSALLAWLTAAAVAIAMLTLTGAAAVAAALHRAYSLGWVGATVGSGLLLL
Rv3630|M.tuberculosis_H37Rv SSALLAWLTAAAVAIAMLTLTGAAAVAAALHRAYSLGWVGATVGSGLLLL
MMAR_5130|M.marinum_M SGALLAWLTAAAVAIAMLTLTGAAAVAAALHRAYSLGWVSATAASGLLLL
MUL_4207|M.ulcerans_Agy99 SGSLLAWLTAAAVAIAMLTLTGAAAVAAALHRAYSLGWVSATAASGLLLL
MAV_0526|M.avium_104 GSALLAWLTAAAVAIAMLTLTGAAAVAAALHRAYALGWVGATVASGLLLA
MSMEG_6140|M.smegmatis_MC2_155 DGTLLAWLTAAAVAIAMLTLTGAAAVAAALHRTYALGWIIATVAAVGLLL
Mflv_1402|M.gilvum_PYR-GCK EGALLAWLTVAAVAIALLTLTGAATVAAALHRAYAAGWVIATAAAALLLM
Mvan_5388|M.vanbaalenii_PYR-1 DGTLLGWLTAAAVAIALLTLTGAATVAAALHRAYAAGWVIATVAATLLLL
TH_0619|M.thermoresistible__bu DGSVLAWLTAAATSIAVLTVTGAATVAAALHRAYAVGWVSATVASTLLLL
MAB_0508c|M.abscessus_ATCC_199 GGPLLGWLTAGAVMIALLTLTGAATVAHARHRAYSAGWVGATVVSALLLL
..:*.***..*. **:**:****:** * **:*: **: **. : **
Mb3654|M.bovis_AF2122/97 LPLSLETRTVVALLCGPLVGIGVHLVALARTDE---------------
Rv3630|M.tuberculosis_H37Rv LPLSLETRTVVALLCGPLVGIGVHLVALARTDE---------------
MMAR_5130|M.marinum_M LPLPLEARTVVALLCGPLVGIGVHLLALARAGYASR------------
MUL_4207|M.ulcerans_Agy99 LPLPLEARTVVALLCGPLVGIGVHLLALARAGYASR------------
MAV_0526|M.avium_104 LPLSLQTRTVVGLLCGPLVGIGVHLVALSRAARLTG------------
MSMEG_6140|M.smegmatis_MC2_155 LPLELSTRTVVALLCGPLVGIAVHLVGLARFSRVSG------------
Mflv_1402|M.gilvum_PYR-GCK LPLDLEVRAVVALLCGPLLGIAVHLSALARDRDGVAPTYAAIDD----
Mvan_5388|M.vanbaalenii_PYR-1 LPLDLEVRTVVALLCGPLLGIAVHLSALARARSIRWHTPAPSDTKETP
TH_0619|M.thermoresistible__bu VPSALETRAVVALLCGPLVGIGVHLLALLGHGPDRRAGTAANTLPT--
MAB_0508c|M.abscessus_ATCC_199 LPLGLEERTVIALLGGPVVGIVVHLIALAFWRDPQLAEDIFD------
:* *. *:*:.** **::** *** .*