For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLMSMATVFTKIINRELPGRFVYEDDDIVAFLTIEPMTQGHTLVVPRAELDNWQDIEPAVFARVMEVSQL IGKAVCKAFDTERSGLIIAGLEVPHLHVHVFPARNLSDFGFANVDRNPSPESLDEAQAKIKAALADLQSA AG
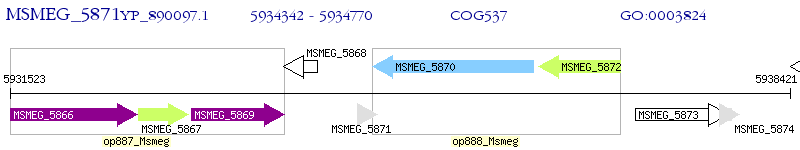
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5871 | - | - | 100% (142) | HIT family protein |
| M. smegmatis MC2 155 | MSMEG_5028 | - | 5e-14 | 33.10% (145) | HIT family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0782c | - | 2e-50 | 82.57% (109) | hypothetical protein Mb0782c |
| M. gilvum PYR-GCK | Mflv_1583 | - | 1e-66 | 88.81% (134) | histidine triad (HIT) protein |
| M. tuberculosis H37Rv | Rv0759c | - | 2e-50 | 82.57% (109) | hypothetical protein Rv0759c |
| M. leprae Br4923 | MLBr_02237 | - | 7e-60 | 78.20% (133) | hypothetical protein MLBr_02237 |
| M. abscessus ATCC 19977 | MAB_0676c | - | 3e-52 | 72.93% (133) | HIT-like (histidine triad) protein |
| M. marinum M | MMAR_4940 | - | 2e-63 | 84.21% (133) | hypothetical protein MMAR_4940 |
| M. avium 104 | MAV_0702 | - | 5e-65 | 88.72% (133) | HIT domain-containing protein |
| M. thermoresistible (build 8) | TH_1250 | - | 4e-63 | 85.71% (133) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0465 | - | 6e-64 | 84.96% (133) | hypothetical protein MUL_0465 |
| M. vanbaalenii PYR-1 | Mvan_5173 | - | 3e-70 | 91.97% (137) | histidine triad (HIT) protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0782c|M.bovis_AF2122/97 ----------------------------MAFLTIEPMTQGHTLVVPRAEI
Rv0759c|M.tuberculosis_H37Rv ----------------------------MAFLTIEPMTQGHTLVVPRAEI
MLBr_02237|M.leprae_Br4923 ----MATIFTKIINRELPGRFVYEDDDVVAFLTIEPMTQGHTLVVPCAEI
MMAR_4940|M.marinum_M ----MASIFTKIINRELPGRFVYEDDDVVAFLTIEPMTQGHTLVVPRAEI
MUL_0465|M.ulcerans_Agy99 ----MASIFTKIINRELPGRFVYEDDDVVAFLTIEPMTQGHTLVVPRAEI
MAV_0702|M.avium_104 ----MASIFTKIINRELPGRFVYEDDDVVAFLTIEPMTQGHTLVVPREEI
MSMEG_5871|M.smegmatis_MC2_155 MLMSMATVFTKIINRELPGRFVYEDDDIVAFLTIEPMTQGHTLVVPRAEL
Mflv_1583|M.gilvum_PYR-GCK ----MATIFTKIINGEIPGRFVYSDDEIVAFLTIEPMTQGHTLVVPRAEI
Mvan_5173|M.vanbaalenii_PYR-1 MLDGMATVFTRIINRELPGRFVYEDDDVVAFLTIEPMTQGHTLVVPREEI
TH_1250|M.thermoresistible__bu ----MATVFTKIINRELPGRFVYEDDEVVAFLTIEPMTQGHTLVVPREEI
MAB_0676c|M.abscessus_ATCC_199 ----MSSVFTKIINGELPGRFVYEDDDVVAFLTIAPITQGHTLVVPRAEI
:***** *:********* *:
Mb0782c|M.bovis_AF2122/97 DHWQNVDPALFGRVMSVSQLIGKAVCRAFSTQRAGMIIAGLEVPHLHIHV
Rv0759c|M.tuberculosis_H37Rv DHWQNVDPALFGRVMSVSQLIGKAVCRAFSTQRAGMIIAGLEVPHLHIHV
MLBr_02237|M.leprae_Br4923 DQWQNVDPAIFGRVIAVSQLIGKGVCRAFNAERAGVIIAGFEVPHLHIHV
MMAR_4940|M.marinum_M DQWQTVDGAVFGRVMEVSQLIGKAVCKAFGTERAGVIIAGLEVPHLHIHV
MUL_0465|M.ulcerans_Agy99 DQWQTVDGAVFGRVMEVSQLIGKAVCKAFGTERAGVIIAGLEVPHLHIHV
MAV_0702|M.avium_104 DNWQDVGSAAFNRVMGVSQLIGKAVCKAFRTERSGLIIAGLEVPHLHVHV
MSMEG_5871|M.smegmatis_MC2_155 DNWQDIEPAVFARVMEVSQLIGKAVCKAFDTERSGLIIAGLEVPHLHVHV
Mflv_1583|M.gilvum_PYR-GCK DNWQDIEPAVFGRVMEVAQLIGKAVCKAFDTERSGVIIAGLEVPHLHVHV
Mvan_5173|M.vanbaalenii_PYR-1 DNWQDIDPAEFARVMEVSQLIGKAVCKAFDTERSGVIIAGLEVPHLHVHV
TH_1250|M.thermoresistible__bu DHWQNIEPEKFAKVMGVAQLIGKAVTKAFDAERAGVIIAGLEVPHLHVHV
MAB_0676c|M.abscessus_ATCC_199 DQWQDIDEALFAKVNLVAQRIGRAVRKAFDAPRAGYIIAGMEVPHLHVHV
*:** : * :* *:* **:.* :** : *:* ****:******:**
Mb0782c|M.bovis_AF2122/97 FPTRSLSDFGFANVDRNPSPGSLDEAQAKIRAALAQLA----
Rv0759c|M.tuberculosis_H37Rv FPTRSLSDFGFANVDRNPSPGSLDEAQAKIRAALAQLA----
MLBr_02237|M.leprae_Br4923 FPTHSLSNFSFANVDRNPSPESLDAAQDKIKAALTQLA----
MMAR_4940|M.marinum_M FPTRHLSDFGFATVDRNPSPQSLDEAQAKIKKALSELT----
MUL_0465|M.ulcerans_Agy99 FPTRHLSDFGFATVDRNPSPESLDEAQAKIKKALSELT----
MAV_0702|M.avium_104 FPTRSLSDFGFANVDRNPSPESLDEAQAKIKAALAQLA----
MSMEG_5871|M.smegmatis_MC2_155 FPARNLSDFGFANVDRNPSPESLDEAQAKIKAALADLQSAAG
Mflv_1583|M.gilvum_PYR-GCK FPARELSDFGFAHVDRNPSPESLDEAQSKIKEALAQLR----
Mvan_5173|M.vanbaalenii_PYR-1 FPARNLSDFGFANVDRNPSPESLDEAQARIKAALAELS----
TH_1250|M.thermoresistible__bu FPAYNLTDFGFANVDRNPSPESLDEAQQKIKAALAELI----
MAB_0676c|M.abscessus_ATCC_199 FPGYQLTDFSFENADPNASAESLDEAQAKIKAALTQID----
** *::*.* .* *.*. *** ** :*: **:::