For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSSPRTRSGIRVSRSSAELGAKSIGTTMHATASSTAHGSPGSNHSPSSTDSASPTGIAMNSIRNAALGWA PRKPPRTSVASTNSSTARMSSAICSALSVFSVTPRWSFIVKLMAMPVSTSTIGAVSRRRCISRDSRDHTA STTEIAISDSNTRKSLQPSHRFPRWRRGYHRQMSQTVAEYVLQRLREWNVEQVFGYPGDGINGLIAAFGK DESAGPRFVQTRHEEMAALAATGYAKFSGNVGVCIATSGPGAIHLLNGLYDAKLDHTPVVAIVGQTARSA MGGSYQQEVDLQALFSDVASDYLVEVNVAGQLPNALDRAFRTARARRAPTAVVIPSDLQEQPYEPPGHAF KQVPSSAPHDAEPVLSPDPAALQDAADVLNAGSKVAILIGQGARGAADEVTQVTDLLGAGVAKALLGKDV LPDDLPFVTGSIGLLGTRPSYELMRDCDTLLIVGTNFPYSQFLPDYGQARAVQIDIDGANIGMRYPTEVN IVAVAKTALAQLVPLLERKDDRSWQQSLEKGVAQWWETVERQSMLKADPVNPMRVVWELSSRLPEYAMVA GDSGSSTNWYARCLKFTSTTRGSLSGTLATMGSAVPYAIGAKFAQPDRPVIALVGDGAMQMNGLAELLTI RRYAHLWADPRMVICVFHNNDLAHVTWELRAMGGAPKFEGSQSLPEVSYADVARVMGLRCITVDDPEQIG AAWDDALSADEPVLLDVHTDPEVPPIPPHVTYEQMKATAQAFVKDPNGWHLFAEIAKNKAAELLPNRR
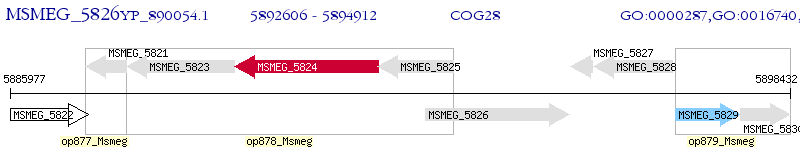
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5826 | - | - | 100% (768) | pyruvate decarboxylase |
| M. smegmatis MC2 155 | MSMEG_3964 | - | 1e-78 | 35.74% (568) | pyruvate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2280 | - | 2e-76 | 35.21% (568) | pyruvate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3028c | ilvB1 | 2e-45 | 30.23% (483) | acetolactate synthase 1 catalytic subunit |
| M. gilvum PYR-GCK | Mflv_3445 | - | 0.0 | 70.78% (592) | pyruvate decarboxylase |
| M. tuberculosis H37Rv | Rv3003c | ilvB1 | 2e-45 | 30.23% (483) | acetolactate synthase 1 catalytic subunit |
| M. leprae Br4923 | MLBr_01696 | ilvB | 8e-48 | 31.47% (464) | acetolactate synthase 1 catalytic subunit |
| M. abscessus ATCC 19977 | MAB_2537c | - | 0.0 | 64.62% (571) | putative pyruvate decarboxylase |
| M. marinum M | MMAR_1710 | ilvB1 | 5e-47 | 30.03% (576) | acetolactate synthase (large subunit) IlvB1 |
| M. avium 104 | MAV_4093 | - | 0.0 | 63.87% (595) | pyruvate decarboxylase |
| M. thermoresistible (build 8) | TH_2440 | - | 3e-50 | 30.14% (584) | PUTATIVE acetolactate synthase, large subunit |
| M. ulcerans Agy99 | MUL_1947 | ilvB1 | 7e-47 | 29.86% (576) | acetolactate synthase 1 catalytic subunit |
| M. vanbaalenii PYR-1 | Mvan_2122 | - | 8e-47 | 29.22% (510) | acetolactate synthase 1 catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1710|M.marinum_M --------------------------------------------------
MUL_1947|M.ulcerans_Agy99 --------------------------------------------------
Mb3028c|M.bovis_AF2122/97 --------------------------------------------------
Rv3003c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01696|M.leprae_Br4923 --------------------------------------------------
TH_2440|M.thermoresistible__bu --------------------------------------------------
Mvan_2122|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5826|M.smegmatis_MC2_155 MSSPRTRSGIRVSRSSAELGAKSIGTTMHATASSTAHGSPGSNHSPSSTD
Mflv_3445|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2537c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4093|M.avium_104 --------------------------------------------------
MMAR_1710|M.marinum_M --------------------------------------------------
MUL_1947|M.ulcerans_Agy99 --------------------------------------------------
Mb3028c|M.bovis_AF2122/97 --------------------------------------------------
Rv3003c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01696|M.leprae_Br4923 --------------------------------------------------
TH_2440|M.thermoresistible__bu --------------------------------------------------
Mvan_2122|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5826|M.smegmatis_MC2_155 SASPTGIAMNSIRNAALGWAPRKPPRTSVASTNSSTARMSSAICSALSVF
Mflv_3445|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2537c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4093|M.avium_104 --------------------------------------------------
MMAR_1710|M.marinum_M -----------------------------MSAPTKRHSPTS----EPQPR
MUL_1947|M.ulcerans_Agy99 -----------------------------MSAPTKRHSPTS----EPQPR
Mb3028c|M.bovis_AF2122/97 -----------------------------MSAPTKPHSPTF----KPEPH
Rv3003c|M.tuberculosis_H37Rv -----------------------------MSAPTKPHSPTF----KPEPH
MLBr_01696|M.leprae_Br4923 -----------------------------MSAPTKPHARPQG-AGNSVPN
TH_2440|M.thermoresistible__bu -----------------------------VSAPTTRTAERAAPHANGAAG
Mvan_2122|M.vanbaalenii_PYR-1 -----------------------------MSAPTKRPPEQSATPTDGTA-
MSMEG_5826|M.smegmatis_MC2_155 SVTPRWSFIVKLMAMPVSTSTIGAVSRRRCISRDSRDHTASTTEIAISDS
Mflv_3445|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2537c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4093|M.avium_104 --------------------------------------------------
MMAR_1710|M.marinum_M GVAAGVNSSP-----VHSKHVAPEQLTGAQSVIRSLEELDVEIIFGIPGG
MUL_1947|M.ulcerans_Agy99 GVAAGVNSSL-----VHSKHVAPEQLTGAQSVIRSLEELDVEIIFGIPGG
Mb3028c|M.bovis_AF2122/97 SAANEPKHPA-----ARPKHVALQQLTGAQAVIRSLEELGVDVIFGIPGG
Rv3003c|M.tuberculosis_H37Rv SAANEPKHPA-----ARPKHVALQQLTGAQAVIRSLEELGVDVIFGIPGG
MLBr_01696|M.leprae_Br4923 TVKPATQFPSK-PAVAKLERVSPEQLTGAQSVIRSLEELDVEVIFGIPGG
TH_2440|M.thermoresistible__bu TAAPEPTNPPAKPASAGQKRIAPHQLTGAQAVIRALEELDVDVIFGIPGG
Mvan_2122|M.vanbaalenii_PYR-1 AAAKHNSAPAPSQATRQPNNIAPQQLTGAQAVIRSLEELGVDVIFGIPGG
MSMEG_5826|M.smegmatis_MC2_155 NTRKSLQPSHRFPRWRRGYHRQMS-QTVAEYVLQRLREWNVEQVFGYPGD
Mflv_3445|M.gilvum_PYR-GCK --------------------MTMS-TDVADFILDRLRQWNVTHIFGYPGD
MAB_2537c|M.abscessus_ATCC_199 ----------------------MS-STVGDILLSRLREWGVQQVFGYPGD
MAV_4093|M.avium_104 ----------------------MSKQEVSDYLLERLRAWGVEHVFGYPGD
.: :: *. .* :** **.
MMAR_1710|M.marinum_M AVLPVYDPLFD--SQKLRHVLVRHEQGAGHAASGYAHATGKVGVCMATSG
MUL_1947|M.ulcerans_Agy99 AVLPVYDPLFD--SQKLRHVLVRHEQGAGHAASGYAHATGKVGVCMATSG
Mb3028c|M.bovis_AF2122/97 AVLPVYDPLFD--SKKLRHVLVRHEQGAGHAASGYAHVTGRVGVCMATSG
Rv3003c|M.tuberculosis_H37Rv AVLPVYDPLFD--SKKLRHVLVRHEQGAGHAASGYAHVTGRVGVCMATSG
MLBr_01696|M.leprae_Br4923 AVLLVYDPLFY--SKKLRHVLVRHEQGAGHAASGYAHVTGKVGVCMATSG
TH_2440|M.thermoresistible__bu AVLPVYDPLFD--SKKLRHVLVRHEQGAGHAASGYAHATGRVGVCMATSG
Mvan_2122|M.vanbaalenii_PYR-1 AVLPVYDPLFD--SKKLRHVLVRHEQGGGHAASGYAHATGRVGVCMATSG
MSMEG_5826|M.smegmatis_MC2_155 GINGLIAAFGKDESAGPRFVQTRHEEMAALAATGYAKFSGNVGVCIATSG
Mflv_3445|M.gilvum_PYR-GCK GINGLVSALGRADDD-PMFVQTRHEEMAAFAAVGYAKFSGEVGVCLATSG
MAB_2537c|M.abscessus_ATCC_199 GINGLLAAWRCADND-PQFVQARHEEMAAFEAVGFAKFSGQLGVCTATSG
MAV_4093|M.avium_104 GINGLLAAWGRADNK-PQFIQSRHEEMSAFQAVGYAKFTGRVGVCAATSG
.: : . . .: ***: .. * *:*: :*.:*** ****
MMAR_1710|M.marinum_M PGATNLVTPLADAQMDSIPVVAITGQVGRGLIGTDAFQEADISGITMPIT
MUL_1947|M.ulcerans_Agy99 PGATNLVTPLADAQMDSIPVVAITGQVGRGLIGTDAFQEADISGITMPIT
Mb3028c|M.bovis_AF2122/97 PGATNLVTPLADAQMDSIPVVAITGQVGRGLIGTDAFQEADISGITMPIT
Rv3003c|M.tuberculosis_H37Rv PGATNLVTPLADAQMDSIPVVAITGQVGRGLIGTDAFQEADISGITMPIT
MLBr_01696|M.leprae_Br4923 PGATNLVTPLADAQMDSVPVVAITGQVGRSLIGTDAFQEADISGITMPIT
TH_2440|M.thermoresistible__bu PGATNLVTPLADAHMDSIPVVAITGQVGRGLIGTDAFQEADISGITMPIT
Mvan_2122|M.vanbaalenii_PYR-1 PGATNLVTPLADAYMDSIPMVAITGQVGRALIGTDAFQEADISGITMPVT
MSMEG_5826|M.smegmatis_MC2_155 PGAIHLLNGLYDAKLDHTPVVAIVGQTARSAMGGSYQQEVDLQALFSDVA
Mflv_3445|M.gilvum_PYR-GCK PGAIHLLNGLYDAKLDHVPVVAIIGQTARTAMGGSYQQEVDLQALFGDVA
MAB_2537c|M.abscessus_ATCC_199 PGAIHLLNGLYDAKLDHVPVLAIVGQADRSAMGGSYQQEVDLLSLYKDVC
MAV_4093|M.avium_104 PGAIHLLNGLYDAKLDHVPVVAIVGQTNRTAMGGNYQQEVDLLSLFKDVA
*** :*:. * ** :* *::** **. * :* . **.*: .: :
MMAR_1710|M.marinum_M -KHNFLVRTGDDIPRVLAEAFHIAASGRPGAVLVDIPKDVLQA-----QC
MUL_1947|M.ulcerans_Agy99 -KHNFLVRTGDDIPRVLAEAFHIAASGRPGAVLVDIPKDVLQA-----QC
Mb3028c|M.bovis_AF2122/97 -KHNFLVRSGDDIPRVLAEAFHIAASGRPGAVLVDIPKDVLQG-----QC
Rv3003c|M.tuberculosis_H37Rv -KHNFLVRSGDDIPRVLAEAFHIAASGRPGAVLVDIPKDVLQG-----QC
MLBr_01696|M.leprae_Br4923 -KHNFLVRAGDDIPRVLAEAFHIASSGRPGAVLVDIPKDVLQG-----QC
TH_2440|M.thermoresistible__bu -KHNFLVRDGDDIPRILAEAFHIASSGRPGPVLVDIPKDILQG-----QC
Mvan_2122|M.vanbaalenii_PYR-1 -KHNFLVRDGDDIARVIAEAFHIASSGRPGPVLVDIPKDILQG-----EC
MSMEG_5826|M.smegmatis_MC2_155 SDYLVEVNVAGQLPNALDRAFRTARARR-APTAVVIPSDLQEQPYEPPGH
Mflv_3445|M.gilvum_PYR-GCK -VYVVEINASAQLPFALDRAIRTARSRR-APTVVIVPSDLQEEEYTPPEH
MAB_2537c|M.abscessus_ATCC_199 SDYVQQCMVPQQLPNLVDRAIRIALSEA-TPTAIIVPSDVFNLGYEAPVH
MAV_4093|M.avium_104 SDYVQMVTVPEQLPNVLDRAIRIAMTQR-APTALIIPADVQELPYSPPEH
: ::. : .*:: * : .. : :* *: :
MMAR_1710|M.marinum_M TFSWPPQMDLPGYKPNTKPHNRQIREAAKLIGAARKPVLYVGGGVIRGEA
MUL_1947|M.ulcerans_Agy99 TFSWPPQMDLPGYKPNTKPHNRQIREAAKLIGAARKPVLYVGGGVIRGEA
Mb3028c|M.bovis_AF2122/97 TFSWPPRMELPGYKPNTKPHSRQVREAAKLIAAARKPVLYVGGGVIRGEA
Rv3003c|M.tuberculosis_H37Rv TFSWPPRMELPGYKPNTKPHSRQVREAAKLIAAARKPVLYVGGGVIRGEA
MLBr_01696|M.leprae_Br4923 KFSWPPKMDLPGYKPNTKPHNRQIRAAAKLIADARKPVLYVGGGVIRGEA
TH_2440|M.thermoresistible__bu TFSWPPRFELPGYKPNTKPHSRQIREAAKLIAAARKPVLYVGGGVIRGAA
Mvan_2122|M.vanbaalenii_PYR-1 TFAWPPQFELPGYKPNTKPHNRQIREAAKLIGEASKPVLYVGGGVIRGEA
MSMEG_5826|M.smegmatis_MC2_155 AFKQVPSSAPHDAEPVLSPDPAALQDAADVLNAGSKVAILIGQGARG--A
Mflv_3445|M.gilvum_PYR-GCK QFKHVPSSDPGDVVGVVTPSDAQVRRAAEILNDGERVAILIGQGARG--A
MAB_2537c|M.abscessus_ATCC_199 AFKQVPSS-LGMSGSRSIPSPDALRAAADLLNSGKKVALLVGQGARG--C
MAV_4093|M.avium_104 AFKMVPSS-LGIECPAIAPEDAAIARAAEILNAGKKVAMLVGTGARG--A
* * * : **.:: . : .: :* *. .
MMAR_1710|M.marinum_M TEQLAELAELTGIPVVTTLMARGAFPDSHRQNMGMPGMHGTVAAVAALQR
MUL_1947|M.ulcerans_Agy99 TEQLAELAELTGIPVVTTLMARGAFPDSHRQNMGMPGMHGTVAAVAALQR
Mb3028c|M.bovis_AF2122/97 TEQLRELAELTGIPVVTTLMARGAFPDSHRQNLGMPGMHGTVAAVAALQR
Rv3003c|M.tuberculosis_H37Rv TEQLRELAELTGIPVVTTLMARGAFPDSHRQNLGMPGMHGTVAAVAALQR
MLBr_01696|M.leprae_Br4923 TEQLRDLAELTGIPVVSTLMARGAFPDSHHQNLGMPGMHGTVAAVAALQR
TH_2440|M.thermoresistible__bu GEELRELAELTGIPVVTTLMARGAFPDSHPQNLGMPGMHGTVAAVAAMQK
Mvan_2122|M.vanbaalenii_PYR-1 SEQLLDLAELTGIPVVTTLMARGAFPDSHPQNLGMPGMHGTVAAVAALQR
MSMEG_5826|M.smegmatis_MC2_155 ADEVTQVTDLLGAGVAKALLGKDVLPDDLPFVTGSIGLLGTRPSYELMRD
Mflv_3445|M.gilvum_PYR-GCK HDEIAGVADVTGAGVAKALLGKDVLPDDLPFVTGSIGLLGTRPSYEMMRD
MAB_2537c|M.abscessus_ATCC_199 VAELTALADLLGAGAAKALLGKDVLPDDLPWVTGSIGLLGTTASWHLMRD
MAV_4093|M.avium_104 HQELAEVADLLGAGAAKALLGKDVLSDELPWVTGSIGLLGTRPSYELMRD
:: :::: * ...:*:.:..:.*. * *: ** .: ::
MMAR_1710|M.marinum_M SDLLIALGTRFDDRVTGKLDSFAPEAKVIHADIDPAEIGKNRNADVPIVG
MUL_1947|M.ulcerans_Agy99 SDLLIALGSRFDDRVTGKLDSFAPEAKVIHADIDPAEIGKNRNADVPIVG
Mb3028c|M.bovis_AF2122/97 SDLLIALGTRFDDRVTGKLDSFAPEAKVIHADIDPAEIGKNRHADVPIVG
Rv3003c|M.tuberculosis_H37Rv SDLLIALGTRFDDRVTGKLDSFAPEAKVIHADIDPAEIGKNRHADVPIVG
MLBr_01696|M.leprae_Br4923 SDLLIALGTRFDDRVTGKLDSFAPDAKVIHADIDPAEIGKNRHADVPIVG
TH_2440|M.thermoresistible__bu SDLLIALGTRFDDRVTGQLDSFAPEAKVIHADIDPAEIGKNRHADVPIVG
Mvan_2122|M.vanbaalenii_PYR-1 SDLLIALGTRFDDRVTGKLDSFAPEAKVIHADIDPAEIGKNRHADVPIVG
MSMEG_5826|M.smegmatis_MC2_155 CDTLLIVGTNFP--YSQFLPDYG-QARAVQIDIDGANIGMRYPTEVNIVA
Mflv_3445|M.gilvum_PYR-GCK CDTLLIVGSNFP--YTQFLPEFG-QARAVQIDDDGASIGMRYPTELNIVA
MAB_2537c|M.abscessus_ATCC_199 CDTLLTVGSNFP--YTQFLPDLG-QARAVQIDRSGKWIGMRYPYELNLVG
MAV_4093|M.avium_104 CDTLLTVGSSFP--YTQFLPECG-QCRAVQIDIDGRLIGMRYPYEVNLVA
.* *: :*: * : * . . :.:.:: * . ** . :: :*.
MMAR_1710|M.marinum_M DVQAVITELIAMLRH-----YDIPGNIEMSEWLAYLEGVRATYPLSYGPQ
MUL_1947|M.ulcerans_Agy99 DVQAVITELIAMLRH-----YDIPGNIEMSEWLAYLEGVRAPYPLSYGPQ
Mb3028c|M.bovis_AF2122/97 DVKAVITELIAMLRH-----HHIPGTIEMADWWAYLNGVRKTYPLSYGPQ
Rv3003c|M.tuberculosis_H37Rv DVKAVITELIAMLRH-----HHIPGTIEMADWWAYLNGVRKTYPLSYGPQ
MLBr_01696|M.leprae_Br4923 DVKAVIVELIAMLRH-----YEVPGNIEMTDWWSYLDGVRKTYPLSYSPQ
TH_2440|M.thermoresistible__bu DVKAVIADLLAVLRR-----DGVADSIKIDGWWEYLRGVQQTYPLSYGPQ
Mvan_2122|M.vanbaalenii_PYR-1 DVKAVITDLIEVLRR-----DGATASTKLDNWWEYLRGIESTYPLSYGPQ
MSMEG_5826|M.smegmatis_MC2_155 VAKTALAQLVPLLERKDDRSWQQSLEKGVAQWWETVE--------RQSML
Mflv_3445|M.gilvum_PYR-GCK DAKATLAALQPHLRRRADTAWRDTVEANVRRWWSVLE--------EQSTL
MAB_2537c|M.abscessus_ATCC_199 DAKDTLQALIPLLERKPDRSWREHIEKKTRGWWTTVQ--------RTAMT
MAV_4093|M.avium_104 DAKAALRALIPHLKRKEDRSWREGIEKNVARWWETMD--------KEAMV
.: .: * *.: * : .
MMAR_1710|M.marinum_M SDGSLGPEYVIEKLGEIAGPDAIYVAGVGQHQMWAAQFIKYEKPRTWLNS
MUL_1947|M.ulcerans_Agy99 SDGSLGPEYVIEKLGEIAGPDAIYVAGVGQHQMWAAQFIKYEKPRTWLNS
Mb3028c|M.bovis_AF2122/97 SDGSLSPEYVIEKLGEIAGPDAVFVAGVGQHQMWAAQFIRYEKPRSWLNS
Rv3003c|M.tuberculosis_H37Rv SDGSLSPEYVIEKLGEIAGPDAVFVAGVGQHQMWAAQFIRYEKPRSWLNS
MLBr_01696|M.leprae_Br4923 SDGTLSPEYVIEKLGEIVGPEAVYVAGVGQHQMWAAQFISYEKPRTWLNS
TH_2440|M.thermoresistible__bu SDGSLSPEYVIETLGKMAGPDAIYVAGVGQHQMWAAQFISYEKPQTWINS
Mvan_2122|M.vanbaalenii_PYR-1 SDGSLSPEFVIETLGKIAGSDAVYVAGVGQHQMWAAQFISYENPKTWINS
MSMEG_5826|M.smegmatis_MC2_155 KADPVNPMRVVWELSSRLPEYAMVAGDSGSSTNWYARCLKFTSTTRGSLS
Mflv_3445|M.gilvum_PYR-GCK SANPVNPMRVAWELSNRLPANAIVTADSGSSTTWYARCVRFTEGVRGSVS
MAB_2537c|M.abscessus_ATCC_199 NADPVNPMRIFWELTKRMPPNAIITADSGSSANWYARHLRFTDGVRGSLS
MAV_4093|M.avium_104 GADPINPLRLFSELSPLLPDNAIVTADSGSSANWYARNLRFRGNMRGSLS
..:.* : * *: ... *. * *: : : *
MMAR_1710|M.marinum_M GGLGTMGFAIPAAMGAKIALPDTEVWAIDGDGCFQMT-NQELATCAIEGI
MUL_1947|M.ulcerans_Agy99 GGLGTMGFAIPAAMGAKIALPDTEVWAIDGDGCFQMT-NQELATCAIEGI
Mb3028c|M.bovis_AF2122/97 GGLGTMGFAIPAAMGAKIALPGTEVWAIDGDGCFQMT-NQELATCAVEGI
Rv3003c|M.tuberculosis_H37Rv GGLGTMGFAIPAAMGAKIALPGTEVWAIDGDGCFQMT-NQELATCAVEGI
MLBr_01696|M.leprae_Br4923 GGLGTMGFAIPAAMGAKIARPEAEVWAIDGDGCFQMT-NQELATCAIEGA
TH_2440|M.thermoresistible__bu GGLGTMGFAVPAAMGAKMGRPDAEVWAIDGDGCFQMT-NQELATCAVEGV
Mvan_2122|M.vanbaalenii_PYR-1 GGLGTMGFAVPAAMGAKMGRPEAEVWAIDGDGCFQMT-NQELATCAIEGV
MSMEG_5826|M.smegmatis_MC2_155 GTLATMGSAVPYAIGAKFAQPDRPVIALVGDGAMQMNGLAELLTIRRYAH
Mflv_3445|M.gilvum_PYR-GCK GTLATMGCAVPYALGAKFAHPDRPVIALVGDGAMQMNGLAELLTVARYRR
MAB_2537c|M.abscessus_ATCC_199 GTLATMGPGVPYAIGAKWAHPDRPGIAFVGDGAMQMNGMNELITIARYWR
MAV_4093|M.avium_104 GTLATMGPGVPYGIGAKFGQPDRPVIVFSGDGAMQMNGMAELITVKRYWN
* *.*** .:* .:*** . * .: ***.:**. ** *
MMAR_1710|M.marinum_M PIKVALINNGNLGMVRQWQNLFYEERYSQTDLATHSHRIP--DFVKLAEA
MUL_1947|M.ulcerans_Agy99 PIKVALINNGNLGMVRQWQNLFYEERYSQTDLATHSHRIP--DFVKLAEA
Mb3028c|M.bovis_AF2122/97 PVKVALINNGNLGMVRQWQSLFYAERYSQTDLATHSHRIP--DFVKLAEA
Rv3003c|M.tuberculosis_H37Rv PVKVALINNGNLGMVRQWQSLFYAERYSQTDLATHSHRIP--DFVKLAEA
MLBr_01696|M.leprae_Br4923 PIKVALINNGNLGMVRQWQALFYQERYSQTDLATHSHRIP--DFVKLAEA
TH_2440|M.thermoresistible__bu PIKVALINNGNLGMVRQWQTLFYEERYSQTDLATHTQRIP--DFVKLAEA
Mvan_2122|M.vanbaalenii_PYR-1 PIKVALINNGNLGMVRQWQTLFYEERYSQTDLATHSRRIP--DFVKLSEA
MSMEG_5826|M.smegmatis_MC2_155 LWADPRMVICVFHNNDLAHVTWELRAMGGAPKFEGSQSLPEVSYADVARV
Mflv_3445|M.gilvum_PYR-GCK EWDDLPFVICVFGNGDLNQVTWELRAMGGSPKFEESQSLPAVSYAEVARA
MAB_2537c|M.abscessus_ATCC_199 QWADPRLVVVVLHNNDLNQVTWEMRAMEGSPKFAESQTLPDVDHARFARG
MAV_4093|M.avium_104 EWKDPRLIIAVLHNNDLNQVTWEMRAMAGAPKFAESQSLPDVDFAAFAAG
: : : : . : :: :* ... .:
MMAR_1710|M.marinum_M LGCVGLRCEREEDVADVINQAREISDRPVVIDFIVGADAQVWP-------
MUL_1947|M.ulcerans_Agy99 LGCVGLRCEREEDVADVINQATEISDRPVVIDFIVGADAQVWP-------
Mb3028c|M.bovis_AF2122/97 LGCVGLRCEREEDVVDVINQARAINDCPVVIDFIVGADAQVWP-------
Rv3003c|M.tuberculosis_H37Rv LGCVGLRCEREEDVVDVINQARAINDCPVVIDFIVGADAQVWP-------
MLBr_01696|M.leprae_Br4923 LGCVGLRCECEEDVVDVINQARAINNRPVVIDFIVGADAQVWP-------
TH_2440|M.thermoresistible__bu LGCVGLRCEREEDVADVIAQARAINDRPVVIDFIVGADAQVWP-------
Mvan_2122|M.vanbaalenii_PYR-1 LGCVGLRCERSEDVADVISQARAINDRPVVIEFVVGADAQVWP-------
MSMEG_5826|M.smegmatis_MC2_155 MGLRCITVDDPEQIGAAWDDALSAD-EPVLLDVHTDPEVPPIPPHVTYEQ
Mflv_3445|M.gilvum_PYR-GCK MGVEARTVDSDEDLGAAWDWALSAT-GPTLLDVRCDPEVPPIPPHVTFDQ
MAB_2537c|M.abscessus_ATCC_199 LDLRGDNIEDPAQIAGAWDRALSAD-RPTVLDVRCDPDVPPIPPHATVDE
MAV_4093|M.avium_104 LGLNAMAIKDPDELSDAWRSALSAD-RPTVLDVYTDPDMPPIPPHATWEQ
:. . :: . * *.:::. ..: *
MMAR_1710|M.marinum_M ------MVAAGTSNDEIQAARGIRPLFDDESEGHA----
MUL_1947|M.ulcerans_Agy99 ------MVAAGTSNDEIQAARGIRPLFDDESEGHA----
Mb3028c|M.bovis_AF2122/97 ------MVAAGTSNDEIQAARGIRPLFDDITEGHA----
Rv3003c|M.tuberculosis_H37Rv ------MVAAGTSNDEIQAARGIRPLFDDITEGHA----
MLBr_01696|M.leprae_Br4923 ------MVAAGASNDEIQAARGIRPLFDDESEGHV----
TH_2440|M.thermoresistible__bu ------MVAAGTSNDEIMAARDIRPLFDESDEGHA----
Mvan_2122|M.vanbaalenii_PYR-1 ------MVAAGTSNDEIQAARGIRPLFD-NEEGHA----
MSMEG_5826|M.smegmatis_MC2_155 MKATAQAFVK-DPNGWHLFAEIAKNKAAELLPNRR----
Mflv_3445|M.gilvum_PYR-GCK MLNMARAVAKGDPAARHLTYQTIKAKAAELFTRS-----
MAB_2537c|M.abscessus_ATCC_199 VTSLVGAVVGGDEDTAGFTRQGIKQKLQQYVPLPKRDRP
MAV_4093|M.avium_104 FKSATAAVLSGDEDRTGFVKVGLKTKMQEFLPHKKS---
. :