For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LVMEKMSLTALARQQLAAARTASAGRSAHTVYGGHEHNLRQTLIALVDGTGLDEHESPGEATLQVIEGRV KLANSEASWEGSPGDLLVIPRTRHSLHALADSVVLLTVVKAVGPHA*
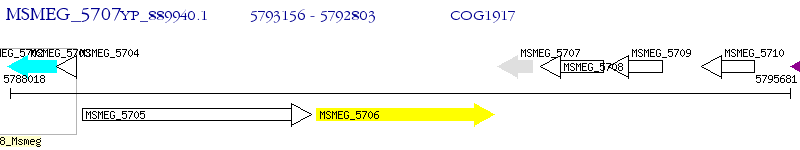
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5707 | - | - | 100% (117) | cupin domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2652c | - | 1e-32 | 55.45% (110) | hypothetical protein Mb2652c |
| M. gilvum PYR-GCK | Mflv_4297 | - | 2e-31 | 60.91% (110) | hypothetical protein Mflv_4297 |
| M. tuberculosis H37Rv | Rv2619c | - | 1e-32 | 55.45% (110) | hypothetical protein Rv2619c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3494c | - | 4e-25 | 53.15% (111) | hypothetical protein MAB_3494c |
| M. marinum M | MMAR_2083 | - | 1e-32 | 55.05% (109) | hypothetical protein MMAR_2083 |
| M. avium 104 | MAV_3497 | - | 2e-31 | 53.64% (110) | cupin domain-containing protein |
| M. thermoresistible (build 8) | TH_2035 | - | 4e-39 | 66.37% (113) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3261 | - | 1e-32 | 55.05% (109) | hypothetical protein MUL_3261 |
| M. vanbaalenii PYR-1 | Mvan_2051 | - | 4e-28 | 56.07% (107) | hypothetical protein Mvan_2051 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4297|M.gilvum_PYR-GCK ---MDVVSLKATAAEQLEAARNAHAGRAAHTVYGGHEHKLRQTAIALLAD
Mvan_2051|M.vanbaalenii_PYR-1 ---MDALSLKSVGDEQLDAARVASAGRAAHTVFGGRDRTLRQTVLALAAG
MSMEG_5707|M.smegmatis_MC2_155 MLVMEKMSLTALARQQLAAARTASAGRSAHTVYGGHEHNLRQTLIALVDG
Mb2652c|M.bovis_AF2122/97 ---MESISLTSLAAEKLAEAQQTHSGRAAHTIHGGHTHELRQTVLALLAG
Rv2619c|M.tuberculosis_H37Rv ---MESISLTSLAAEKLAEAQQTHSGRAAHTIHGGHTHELRQTVLALLAG
MAV_3497|M.avium_104 ---MDSVSLTDLAAEKLAEAKQSHSGRAAHTIHGGHTHELRQTVLALLAD
MMAR_2083|M.marinum_M ---MESLSLTALASEKLAEAKQSHSGRAAHTIHGGHAHELRQTVLALLAD
MUL_3261|M.ulcerans_Agy99 ---MESLSLTALASEKLAEAKQSHSGRAAHTIHGGHAHELRQTVLALLAD
TH_2035|M.thermoresistible__bu ---MEKISLTALARQELASAKAASSGRSAQTIFGGHHRALRQTLMALTAG
MAB_3494c|M.abscessus_ATCC_199 ---MQKRSIDAVIRQLFERADNGHN--AADTVVGGHERILRQTVIALREG
*: *: : : * :*.*: **: : **** :** .
Mflv_4297|M.gilvum_PYR-GCK HRLDDHESPGEASLLVLHGRVRLSTSSESAELGEGEYLIIPAERHNLAAL
Mvan_2051|M.vanbaalenii_PYR-1 NRLDDHESPGEATLLVLRGKVQIGTATDSVDGGEGDYLVIPDEIHNLVAL
MSMEG_5707|M.smegmatis_MC2_155 TGLDEHESPGEATLQVIEGRVKLANSEASWEGSPGDLLVIPRTRHSLHAL
Mb2652c|M.bovis_AF2122/97 HDLSEHDSPGEATLQVLQGHVCLTAGEDAWNGRAGDYVAIPPTRHALHAV
Rv2619c|M.tuberculosis_H37Rv HDLSEHDSPGEATLQVLQGHVCLTAGEDAWNGRAGDYVAIPPTRHALHAV
MAV_3497|M.avium_104 HDLSEHDSPGEATLQVLQGHVRLTTGGDAWEGRAGEYVAIPAERHALHAV
MMAR_2083|M.marinum_M HNLGEHDSPGEATLQVLQGHVRLTTGDDAWDGKTGDYVAIPPQRHALHAI
MUL_3261|M.ulcerans_Agy99 HNLGEHDSPGEATLQVLQGHVRLTTDDDAWDGKTGDYVAIPPQRHALHAI
TH_2035|M.thermoresistible__bu SELAEHDSPGEATLQVLQGRVRLTAGPHGWDGSPGDHLMIPAERHALRAI
MAB_3494c|M.abscessus_ATCC_199 AELGEHESPGEATIYVIRGSVRLVAGAEHWDGRAGDLIEIPDARHSLQAL
* :*:*****:: *:.* * : : *: : ** * * *:
Mflv_4297|M.gilvum_PYR-GCK EDSVVLLTVVTGI-------
Mvan_2051|M.vanbaalenii_PYR-1 EDSVVLLTVVARR-------
MSMEG_5707|M.smegmatis_MC2_155 ADSVVLLTVVKAVGPHA---
Mb2652c|M.bovis_AF2122/97 EDSVIMLTVLKSLPDAHSGS
Rv2619c|M.tuberculosis_H37Rv EDSVIMLTVLKSLPDAHSGS
MAV_3497|M.avium_104 EDSVVLLTVLKTIPSAH---
MMAR_2083|M.marinum_M EDSVVVLTVLKSQPGAH---
MUL_3261|M.ulcerans_Agy99 EDSVVVLTVLKSQPGAH---
TH_2035|M.thermoresistible__bu EDSVVLLTVVKP-VGPHT--
MAB_3494c|M.abscessus_ATCC_199 TDSAVLLTVAKRRG------
**.::***