For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKRTFGILALVVVLAIAGTGIGVWRLSSSHGENLPQISAYSHGHTVRTGPFQYCPVLDLDNCLDPREAAE LRVTEKYPVQLSVPSAIGRAPWRLLRVYENPIDTTVSVYRPNTTLAVTIPTVDPQRGRLTGLAVQLLTLV RDQNGEEFALPHAEWSISTTWS
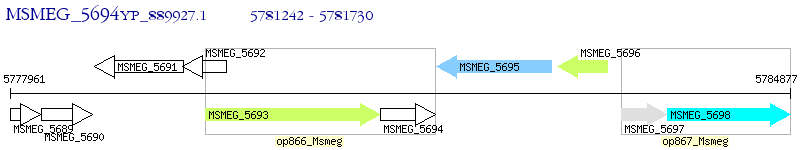
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5694 | - | - | 100% (162) | hypothetical protein MSMEG_5694 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0899c | - | 4e-51 | 58.23% (158) | hypothetical protein Mb0899c |
| M. gilvum PYR-GCK | Mflv_1710 | - | 8e-49 | 55.90% (161) | hypothetical protein Mflv_1710 |
| M. tuberculosis H37Rv | Rv0875c | - | 4e-51 | 58.23% (158) | hypothetical protein Rv0875c |
| M. leprae Br4923 | MLBr_02144 | - | 4e-46 | 53.75% (160) | hypothetical protein MLBr_02144 |
| M. abscessus ATCC 19977 | MAB_0874c | - | 7e-28 | 41.92% (167) | hypothetical protein MAB_0874c |
| M. marinum M | MMAR_4657 | - | 4e-51 | 60.13% (158) | hypothetical protein MMAR_4657 |
| M. avium 104 | MAV_1004 | - | 3e-49 | 56.13% (155) | hypothetical protein MAV_1004 |
| M. thermoresistible (build 8) | TH_1067 | - | 1e-54 | 65.16% (155) | POSSIBLE CONSERVED EXPORTED PROTEIN |
| M. ulcerans Agy99 | MUL_0275 | - | 1e-50 | 59.49% (158) | hypothetical protein MUL_0275 |
| M. vanbaalenii PYR-1 | Mvan_5041 | - | 1e-49 | 57.14% (161) | hypothetical protein Mvan_5041 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5694|M.smegmatis_MC2_155 MK-----RTFGILALVVVLAIAGTGIGVWRLSSSHGENLPQISAYSHGHT
TH_1067|M.thermoresistible__bu -----------MLAAVAVLAAVGTGLLITRLSAQQDHQLPRISVYSAGHL
Mflv_1710|M.gilvum_PYR-GCK MK-----RLVAVLAAVALLASIGTGVLVWQLSRDHGPGSPEISAFSHGNL
Mvan_5041|M.vanbaalenii_PYR-1 MK-----RVVAVLAAVALLSAVGTGVLVWQLMRDHSPELPEISAYSHGHL
MMAR_4657|M.marinum_M MK----RWMTALIAVVVLVLSASAGVGAWLLARNSGPQRPEISLYSHGHL
MUL_0275|M.ulcerans_Agy99 MK----RWMTALIAVVVLGLSASAGVGAWLLARNSGPQRPEISLYSHGHL
Mb0899c|M.bovis_AF2122/97 MK----RGVATLPVILVILLSVAAGAGAWLLVRGHGPQQPEISAYSHGHL
Rv0875c|M.tuberculosis_H37Rv MK----RGVATLPVILVILLSVAAGAGAWLLVRGHGPQQPEISAYSHGHL
MAV_1004|M.avium_104 MKPKTRRGVAVLLACLMIVLCAGAGVGTWLLVGRGGPQHPEISAYSHGHL
MLBr_02144|M.leprae_Br4923 MQ----RRVTVLLPAVVMLLAAAAGFGVWLLVREPDPQRPEISVYSHGHL
MAB_0874c|M.abscessus_ATCC_199 MK----AKLTALAVVLVLTAAAMVGFITWR-VRGHQEERPEISAFTHGTV
: : : .* *.** :: *
MSMEG_5694|M.smegmatis_MC2_155 VRTGPFQYCPVLDLDNCLDPREAAELRVTEKYPVQLSVPSAIGRAPWRLL
TH_1067|M.thermoresistible__bu IRVGPFQYCPVLDLDNCELSGEQGELPVNERDVVQLSVPGAIAGGPWRLL
Mflv_1710|M.gilvum_PYR-GCK TRVGPYRYCQVFNPTDCAVPAEQGELAVDARHPVQLSIPSAISRAPWVLL
Mvan_5041|M.vanbaalenii_PYR-1 TRVGPYRFCEVLNPTDCVVSADQGELPVTPSDPVQLSIPGEIARAPWVLL
MMAR_4657|M.marinum_M TRVGPYMYCDVLRLDECQTPQTQGELPVTERYPVQLSVPQAISRAPWRLL
MUL_0275|M.ulcerans_Agy99 TRVGPYMYCDVLRLDECQTPQTQGELPVTERYPVQLSVPQVISRAPWRLL
Mb0899c|M.bovis_AF2122/97 TRVGPYLYCNVVDLDDCQTPQAQGELPVSERYPVQLSVPEVISRAPWRLL
Rv0875c|M.tuberculosis_H37Rv TRVGPYLYCNVVDLDDCQTPQAQGELPVSERYPVQLSVPEVISRAPWRLL
MAV_1004|M.avium_104 TRVGPYLYCNVLNLEDCETPQSQGELPVSERYPIQLSVPDAIARAPWRLL
MLBr_02144|M.leprae_Br4923 TRVGPYLHCNMLNLDECQTPQAQGVLSADEHNPVQLSVPETISRAPWRLL
MAB_0874c|M.abscessus_ATCC_199 IHAAPYQYCDKNKLDQCDPLGHTVQLAVNAHDPVQLSVDGRIARAPWMLS
:..*: .* :* * . :***: *. .** *
MSMEG_5694|M.smegmatis_MC2_155 RVYENP-----IDTTVSVYRPNTTLAVTIPTVDPQRGRLTGLAVQLLTLV
TH_1067|M.thermoresistible__bu RVYEDP-----IDTTVSHYRPDTRFAETIETVDPHRGRLTGIAVQLMTLV
Mflv_1710|M.gilvum_PYR-GCK RAYTD-------DDIVEIVPPHTRLAVTIPTEDPERGRLTGIAVQLPTWV
Mvan_5041|M.vanbaalenii_PYR-1 RAYED-------ADLVEEFRPGTTLAVTIPTVDAQRGRLTGIAVQLPTLV
MMAR_4657|M.marinum_M QLYDDP-----TNTTAVIFRPNSRLAVTIPTVDPQRGRLTGVVVQLLTLV
MUL_0275|M.ulcerans_Agy99 QLYDDP-----TNTTAVIFRPNSRLAVTIPTVDPQRGRLTGVFVQLLTLV
Mb0899c|M.bovis_AF2122/97 QVYQDP-----ANTTSTLFRPDTRLAVTIPTVDPQRGRLTGIVVQLLTLV
Rv0875c|M.tuberculosis_H37Rv QVYQDP-----ANTTSTLFRPDTRLAVTIPTVDPQRGRLTGIVVQLLTLV
MAV_1004|M.avium_104 QVYEDP-----GNTTSTIYRPGTRFAVTIPSVDPHRGRLAGIVVQLLTLA
MLBr_02144|M.leprae_Br4923 RIYEDP-----TGTTSTLYRPNTRLAVTIPTLDPHRGRLTGIVVQLLTLV
MAB_0874c|M.abscessus_ATCC_199 RIYAPTGDVDHKRAESSVYR-DHRTAVTIPTIDGEGRRLLGLEIKLPTVV
: * * ** : * . ** *: ::* * .
MSMEG_5694|M.smegmatis_MC2_155 RDQNGEEFALPHAEWSISTTWS-----------
TH_1067|M.thermoresistible__bu QDENGELFDVPHAEWSVATVWPD----------
Mflv_1710|M.gilvum_PYR-GCK RDEAGNEFPVPHAEWAVQTVWPDEPAGDPQPAE
Mvan_5041|M.vanbaalenii_PYR-1 RDESGNEFPVPHAEWSVRTVWR---AG------
MMAR_4657|M.marinum_M VDPSGELREAPHAEWSVRVVF------------
MUL_0275|M.ulcerans_Agy99 VDPSGELREAPHAEWSVRVVF------------
Mb0899c|M.bovis_AF2122/97 VDHSGELRDVPHAEWSVRLIF------------
Rv0875c|M.tuberculosis_H37Rv VDHSGELRDVPHAEWSVRLIF------------
MAV_1004|M.avium_104 MDPAGEVRDVPHAEWSVRLTF------------
MLBr_02144|M.leprae_Br4923 VDPAGEVHDVPHTEWLVRLTF------------
MAB_0874c|M.abscessus_ATCC_199 QTETGDLLEVTHAVWSVATVWKD----------
*: .*: * : :