For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPTPHAAEIVLSADERAELEGWARRRTSAAGLAMRARIVLAAADGGSNTELADRLELSIGTVRRWRNRFA ELRLDGLVDEPRPGRPRVVGDEQIEALITATLETTPPDATHWSTRSMAAHLGLSQSMVSRVWRAFGLAPH KQDSWKLSKDPLFVAKVRDVVGLYLNPPERALVLCVDEKTQIQALNRTQPVFPMLPGTPARASHDYVRHG TSSLYAALDLTTGKVIGALHSRHRATEFLAFLKKIDVEVPDDLDVHLVLDNASTHKTPAVKRWLTGHPRF VLHFTPTSSSWLNLVERWFAELTTKKLRRSTHTSVRQLNADIRAWIDTWNDNPKPYVWTKTADQILASIA NYCARINDSGH
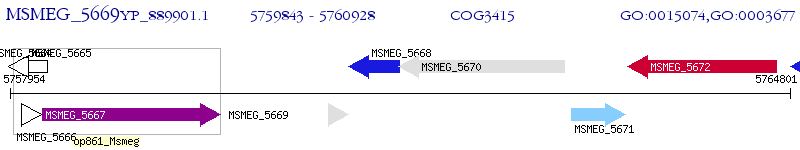
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5669 | - | - | 100% (361) | ISMsm2, transposase |
| M. smegmatis MC2 155 | MSMEG_4522 | - | 0.0 | 100.00% (361) | ISMsm2, transposase |
| M. smegmatis MC2 155 | MSMEG_3510 | - | 0.0 | 100.00% (361) | ISMsm2, transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1425 | - | 6e-12 | 56.36% (55) | transposase for insertion sequence |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5669|M.smegmatis_MC2_155 MPTPHAAEIVLSADERAELEGWARRRTSAAGLAMRARIVLAAADGGSNTE
MMAR_1425|M.marinum_M --------------------------------------------------
MSMEG_5669|M.smegmatis_MC2_155 LADRLELSIGTVRRWRNRFAELRLDGLVDEPRPGRPRVVGDEQIEALITA
MMAR_1425|M.marinum_M --------------------------------------------------
MSMEG_5669|M.smegmatis_MC2_155 TLETTPPDATHWSTRSMAAHLGLSQSMVSRVWRAFGLAPHKQDSWKLSKD
MMAR_1425|M.marinum_M --------------------------------------------------
MSMEG_5669|M.smegmatis_MC2_155 PLFVAKVRDVVGLYLNPPERALVLCVDEKTQIQALNRTQPVFPMLPGTPA
MMAR_1425|M.marinum_M --------------------------------------------------
MSMEG_5669|M.smegmatis_MC2_155 RASHDYVRHGTSSLYAALDLTTGKVIGALHSRHRATEFLAFLKKIDVEVP
MMAR_1425|M.marinum_M --------------------------------------------------
MSMEG_5669|M.smegmatis_MC2_155 DDLDVHLVLDNASTHKTPAVKRWLTGHPRFVLHFTPTSSSWLNLVERWFA
MMAR_1425|M.marinum_M ----MHLILNHDATHKHSAVRVWLDGRPRLHLDAVAASSSWLDLVDRWLR
:**:*:: :*** .**: ** *:**: *. ..:*****:**:**:
MSMEG_5669|M.smegmatis_MC2_155 ELTTKKLRRSTHTSVRQLNADIRAWIDTWNDNPKPYVWTKTADQILASIA
MMAR_1425|M.marinum_M ELTEKALRRVAFHSMP----------------------------------
*** * *** :. *:
MSMEG_5669|M.smegmatis_MC2_155 NYCARINDSGH
MMAR_1425|M.marinum_M -----------