For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTKPEIEFPDGPAPTELVITDLVVGDGPEAVAGANVEVHYVGVEYDTGEEFDSSWNRGESIEFPLRGLIQ GWQDGIPGMRVGGRRKLIIPPEQAYGPAGGGHRLSGKTLIFVIDLLATR*
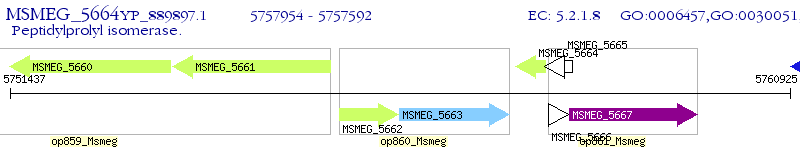
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5664 | - | - | 100% (120) | binding protein |
| M. smegmatis MC2 155 | MSMEG_3434 | - | 2e-12 | 37.84% (111) | peptidyl-prolyl cis-trans isomerase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1740 | - | 1e-62 | 92.44% (119) | peptidylprolyl isomerase, FKBP-type |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0954 | - | 2e-50 | 74.58% (118) | putative peptidyl-prolyl cis-trans isomerase FkbP |
| M. marinum M | MMAR_1374 | - | 1e-33 | 58.33% (120) | FK-506 binding protein, peptidyl-prolyl cis- trans isomerase |
| M. avium 104 | MAV_2725 | - | 3e-12 | 33.90% (118) | peptidyl-prolyl cis-trans isomerase domain-containing protein |
| M. thermoresistible (build 8) | TH_0455 | - | 7e-61 | 91.45% (117) | binding protein |
| M. ulcerans Agy99 | MUL_2502 | - | 8e-33 | 56.67% (120) | FK-506 binding protein, peptidyl-prolyl cis- trans isomerase |
| M. vanbaalenii PYR-1 | Mvan_5011 | - | 4e-63 | 92.50% (120) | peptidylprolyl isomerase, FKBP-type |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1374|M.marinum_M --------------------------------------------------
MUL_2502|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1740|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5011|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5664|M.smegmatis_MC2_155 --------------------------------------------------
TH_0455|M.thermoresistible__bu --------------------------------------------------
MAB_0954|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2725|M.avium_104 MNSVRTFSVAAFAACFTAAAAMLASAGTAGAADSCPTAAPPSGGTPDWTL
MMAR_1374|M.marinum_M ------------MTAKKPEIDFPGGEPPTDLVITDVVEGDGAEATSGKTV
MUL_2502|M.ulcerans_Agy99 ------------MTAKKPEIDFPGGEPPTDLVITDVVEGDGAEATSGKTL
Mflv_1740|M.gilvum_PYR-GCK -------------MATKPEIEFPDGPAPTELVIEDVVVGDGSEAVAGANV
Mvan_5011|M.vanbaalenii_PYR-1 -------------MATKPEIEFPDGPAPAGLVIEDIVVGDGPEAVPGANV
MSMEG_5664|M.smegmatis_MC2_155 -------------MTTKPEIEFPDGPAPTELVITDLVVGDGPEAVAGANV
TH_0455|M.thermoresistible__bu ------VVAVTRNSAQKPEIDFPDGPAPTELVIEDLIVGDGPEAVAGGTV
MAB_0954|M.abscessus_ATCC_1997 --------------MTKPEIDFQPGPPPTELVIKDITVGDGAEATPGSVV
MAV_2725|M.avium_104 TGTTGSVAVTGSTDTAAPVVNVTAPFSVTQTQVHTLRAADGPAVPGTARV
* ::. . : : : .**. . :
MMAR_1374|M.marinum_M VVHYVGVAHSTGEEFDASYNRGDPLMFKLGVGQVIQGWDQGVQGMKVGGR
MUL_2502|M.ulcerans_Agy99 VVHYVGVAHSTGEEFDASYNRGDPLMFKLGVGQVIQGWDQGVQGMKVGGR
Mflv_1740|M.gilvum_PYR-GCK EVHYVGVEYDTGEEFDSSWNRGESIEFPLRG--LIQGWQDGIPGMKVGGR
Mvan_5011|M.vanbaalenii_PYR-1 EVHYVGVEYDTGEEFDSSWNRGESIEFPLRG--LIQGWQDGIPGMKVGGR
MSMEG_5664|M.smegmatis_MC2_155 EVHYVGVEYDTGEEFDSSWNRGESIEFPLRG--LIQGWQDGIPGMRVGGR
TH_0455|M.thermoresistible__bu EVHYVGVEYDTGEEFDSSWNRGESIAFPLRG--LIAGWQEGIPGMRVGGR
MAB_0954|M.abscessus_ATCC_1997 DVHYVGVEFESGEEFDSSWNRGESIEFPLSA--LIKGWQDGIPGMKVGGR
MAV_2725|M.avium_104 SVCYMGVNGRDGTVFDSSYQRGAPVDFPLGG--VVPGFQKAIAGQKVGST
* *:** * **:*::** .: * * :: *::..: * :**.
MMAR_1374|M.marinum_M RQLHIPAHLAYGDRGAGGVIKPGESLIFVVDLLDVR-
MUL_2502|M.ulcerans_Agy99 RQLHIPAHLAYGDRCAGGVIKPGESLIFVVDLLDVR-
Mflv_1740|M.gilvum_PYR-GCK RKLTIPPEQAYGPAGGGHRLS-GKTLIFMIDLLGTS-
Mvan_5011|M.vanbaalenii_PYR-1 RKLTIPPEQAYGPAGGGHRLS-GKTLIFVIDLLATR-
MSMEG_5664|M.smegmatis_MC2_155 RKLIIPPEQAYGPAGGGHRLS-GKTLIFVIDLLATR-
TH_0455|M.thermoresistible__bu RKLTIPPELAYGPAGGGHRLS-GKTLIFVIDLLATR-
MAB_0954|M.abscessus_ATCC_1997 RQLTVPPAQAYGEAGAGHQLS-GKTLVFVIDLLGAR-
MAV_2725|M.avium_104 VAVAMTSADGYPDGQPSAGIRPGDTLVFAIKILGATT
: :.. .* . : *.:*:* :.:* .