For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVTPTILNGGEPASIGFGGGTVTLVEGATFCLSDRHGDVLAGRSHGLFFRDARVLSRWQLHVDGGEPESL TVQTPEAFAAQFIMRRTPRAGVADSTLLLVRDRLIADGLRETISLYNLADEPTVVSLELLVDADFADLFA VKEGRAPIGGAEISLVDNELVLNDRSNRVRGVKITASGSPVVTPGVFRWRVVVPPRGTWQAEVVAEPTWA NQSVKTRFRTGEELEASAPARKIEAWRGTTTTVETDLRALAATLQRSESDLGALLIRDETDERPYVAAGA PWFMTLFGRDSLLTAWMSLPLDVGLSVGTLQRLADAQGRRVDPLTEEQPGRIMHEIRRGPASTDVLGGSV YYGSADSTPLFVMLLAECWRWGADEGAVRALLPAADAALSWAARYGDRDDDGFIEYKRATDRGLLNQGWK DSFDAITDAAGHRADAPIALCEVQAYQYAALRARAELADAFGEPATAKRLRDRAATLKKRFAEKFWLPDR GYYALALDGDKRPVDSLTSNVGHCLWAGIATDEHAARLVEKLGTSEMDSGFGLRTLAADMGAYNPMSYHN GSVWPHDTAIAVAGLLRYRHVPGAVELAERLALGLLDAADAFGGRLPELFCGFHRSQFASPVPYPTSCSP QAWASAAPLLLVRAFLGLEPDVPHRKLTVRPQLPAAWGRVALTDLRLGSATVAVEAEGTVAKVTGLPGDW QLSTAES*
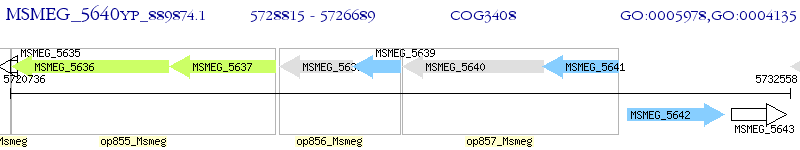
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5640 | - | - | 100% (708) | amylo-alpha-1,6-glucosidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1025 | - | 0.0 | 73.04% (701) | hypothetical protein MAV_1025 |
| M. thermoresistible (build 8) | TH_0093 | - | 0.0 | 72.90% (701) | amylo-alpha-1,6-glucosidase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1025|M.avium_104 -MSVATAFNGGAPAQIGSGGDAVTLVEGGTFCLSNRHGDVPVGTSYGLFF
TH_0093|M.thermoresistible__bu ---MTNALNAGNVAAIGSGRDTVTLVEGGTFCLSDRHGDVLEG-AHGLFF
MSMEG_5640|M.smegmatis_MC2_155 MTVTPTILNGGEPASIGFGGGTVTLVEGATFCLSDRHGDVLAGRSHGLFF
.. :*.* * ** * .:******.*****:***** * ::****
MAV_1025|M.avium_104 RDARVLSRWELRVDGQVAEALSVEATEAFAAQFILRRAPRSGLADSTLLI
TH_0093|M.thermoresistible__bu RDARALSRWELRVDGRSAEPLTVLTPEAFAAQFVLRRTPRPGLADSTLLL
MSMEG_5640|M.smegmatis_MC2_155 RDARVLSRWQLHVDGGEPESLTVQTPEAFAAQFIMRRTPRAGVADSTLLL
****.****:*:*** .*.*:* :.*******::**:**.*:******:
MAV_1025|M.avium_104 VRERLVADGLRETISLHNLDKESTVVSLELHADADFADLFAVKEGRAATA
TH_0093|M.thermoresistible__bu VRERLIADGMRETITLHNLDDEPTVVSLELFVDGDFVDLFAVKEGRVVPG
MSMEG_5640|M.smegmatis_MC2_155 VRDRLIADGLRETISLYNLADEPTVVSLELLVDADFADLFAVKEGRAPIG
**:**:***:****:*:** .*.******* .*.**.*********. .
MAV_1025|M.avium_104 GADMHVADGELVLRGRADQVRGLAVSASGDPIVLPGSLNWRVVVPPGGRW
TH_0093|M.thermoresistible__bu GAEMTVVDDELVLRDRGDLVRGLTITATGVDKVLPGTLRWRVVVPPRSSW
MSMEG_5640|M.smegmatis_MC2_155 GAEISLVDNELVLNDRSNRVRGVKITASGSPVVTPGVFRWRVVVPPRGTW
**:: :.*.****..*.: ***: ::*:* * ** :.******* . *
MAV_1025|M.avium_104 QTEVMVQPTWSNRKVRTRIPRGKDIESSAPARKIERWRDTATTVETDHAG
TH_0093|M.thermoresistible__bu QTEIVAQPTWANQQVHSRFRSGERLDASAPAQKISAWRDVTTTVECDHPT
MSMEG_5640|M.smegmatis_MC2_155 QAEVVAEPTWANQSVKTRFRTGEELEASAPARKIEAWRGTTTTVETDLRA
*:*::.:***:*:.*::*: *: :::****:**. **..:**** *
MAV_1025|M.avium_104 LAQVLRQTESDLGALLIHDADGRGRPYVAAGAPWFMTLFGRDSLLTAWMT
TH_0093|M.thermoresistible__bu LAEVLQRTESDLGALLVYD-DGDGRPFVAAGAPWYMTLFGRDSLLTAWMA
MSMEG_5640|M.smegmatis_MC2_155 LAATLQRSESDLGALLIRD-ETDERPYVAAGAPWFMTLFGRDSLLTAWMS
** .*:::********: * : **:*******:**************:
MAV_1025|M.avium_104 LPLDVGLSIGTLQRLAALQGRRVDALTEEEPGRIMHEIRRGPASADVLGG
TH_0093|M.thermoresistible__bu LPIDVDLSIGTLKQLAELQGRQINPITEEEPGRIMHEIRRGPASADALGG
MSMEG_5640|M.smegmatis_MC2_155 LPLDVGLSVGTLQRLADAQGRRVDPLTEEQPGRIMHEIRRGPASTDVLGG
**:**.**:***::** ***:::.:***:**************:*.***
MAV_1025|M.avium_104 SVYYGTADATPLFVMLLAESWQWGADEAVIRSLLPAADAALAWAEQYGDR
TH_0093|M.thermoresistible__bu NVYYGSVDASLLFVMLLGEAWRWGADEDAVRSLLPAADAALQWAERYGDR
MSMEG_5640|M.smegmatis_MC2_155 SVYYGSADSTPLFVMLLAECWRWGADEGAVRALLPAADAALSWAARYGDR
.****:.*:: ******.*.*:***** .:*:********* ** :****
MAV_1025|M.avium_104 DGDGFVEYRRATDRGLINQGWKDSFNGINDAKGRVAEPPIALCEVQGYVY
TH_0093|M.thermoresistible__bu DGDGFVEYRRATDRGLINQGWKDSYDGINHASGRLAEPPIALCEVQGYHY
MSMEG_5640|M.smegmatis_MC2_155 DDDGFIEYKRATDRGLLNQGWKDSFDAITDAAGHRADAPIALCEVQAYQY
*.***:**:*******:*******::.*..* *: *:.********.* *
MAV_1025|M.avium_104 AALLARAELAEGMGELAQAAQLRERAQALRTRFGEAFWLPDRGWYAVALD
TH_0093|M.thermoresistible__bu AALLARAELADAFGEPALAELLRGRAEALRIGFQERFWMPDKGYYAVALD
MSMEG_5640|M.smegmatis_MC2_155 AALRARAELADAFGEPATAKRLRDRAATLKKRFAEKFWLPDRGYYALALD
*** ******:.:** * * ** ** :*: * * **:**:*:**:***
MAV_1025|M.avium_104 GRKDRVDALTSNVGHCLWTGIATDEHAAAIVERLAGEAMDSGFGLRTLAT
TH_0093|M.thermoresistible__bu RDKRQVDALTSNVGHCLWTGIASDQHAERIIENLSGHQMDSGFGLRTLAT
MSMEG_5640|M.smegmatis_MC2_155 GDKRPVDSLTSNVGHCLWAGIATDEHAARLVEKLGTSEMDSGFGLRTLAA
* **:**********:***:*:** ::*.*. ***********:
MAV_1025|M.avium_104 TMGAYNPMSYHNGSVWPHDTAITVSGLLRYGHIPGALALAERLATGLLDA
TH_0093|M.thermoresistible__bu TMGAYNPMSYHNGSVWPHDTAIAVAGLLRYRHVRGAVELAERLTAGLLDA
MSMEG_5640|M.smegmatis_MC2_155 DMGAYNPMSYHNGSVWPHDTAIAVAGLLRYRHVPGAVELAERLALGLLDA
*********************:*:***** *: **: *****: *****
MAV_1025|M.avium_104 AAAFGGRLPELFCGFPRSQFASPVPYPTSCSPQAWASAAPLLLLRSFLGL
TH_0093|M.thermoresistible__bu AAAFGGRLPELYCGFPREQFPAPVPYPTSCSPQAWASAAPLLLVRSFLGF
MSMEG_5640|M.smegmatis_MC2_155 ADAFGGRLPELFCGFHRSQFASPVPYPTSCSPQAWASAAPLLLVRAFLGL
* *********:*** *.**.:*********************:*:***:
MAV_1025|M.avium_104 DPHVPNRALTVTPHLPAEWGRIALSNLRLGATTVQLEAEGETVKVQGLGD
TH_0093|M.thermoresistible__bu EPHVPERRLTVHPRLPSSWGRLTLAELRLGSRSVNIEAHCDTVKVDGLPE
MSMEG_5640|M.smegmatis_MC2_155 EPDVPHRKLTVRPQLPAAWGRVALTDLRLGSATVAVEAEGTVAKVTGLPG
:*.**.* *** *:**: ***::*::****: :* :**. ..** **
MAV_1025|M.avium_104 DWQLVTP---------
TH_0093|M.thermoresistible__bu DWELVTSSADESPPLG
MSMEG_5640|M.smegmatis_MC2_155 DWQLSTAES-------
**:* *.