For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVRPLVTTGAALISAGALVAATPALFVPKDEIAIAAPSMATAPKQLTVDQVDLLALSLQGLYQAFFQGYG GYANPGTEEVTTYERNPDGTLVLDANGDPIPITTEEPVFGDCTATGAVCLSGFPGAAYYLSDELLPLGDV DNIFFESGIEPFFQIAAQTIAAEVDALDPTGRLTLERRVADFFEGGATQLVGNTLLDNLSEGTVAYGLTN SFFFGYGENAGVVAALTYVVDAIAQGTADPVPNPINPLGPGVSSATVETQLVSKQAIEQKVVEGEDPAST NILTRSRFSVPVSGEALGFKSDANTTPVLEKDGLGEETTSDGVAPAVNVTSPQSAALGQAPETPATAPEE PSAPAAEVKEETESTGATANTKDGNKFTPGVILSPGGRKSSSANGWEKLEQDVKKGINTLGKIFNKGKTS KPATSSTTDNAGEGGEGGGDE*
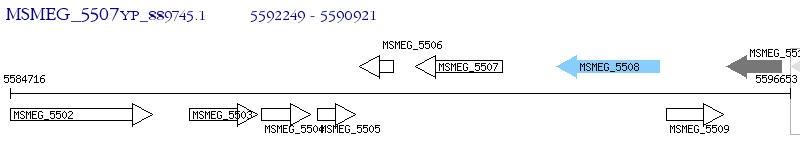
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5507 | - | - | 100% (442) | hypothetical protein MSMEG_5507 |
| M. smegmatis MC2 155 | MSMEG_5501 | - | 7e-06 | 24.89% (237) | hypothetical protein MSMEG_5501 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_4419 | - | 2e-22 | 27.83% (424) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5507|M.smegmatis_MC2_155 MAVRPLVTTGAALISAGALVAATPALFVPKDEIAIAAPSMATAPKQLTVD
TH_4419|M.thermoresistible__bu MAVRPLIGAGVVLSSAGLLIASLPTIAPPLLDREIRIAADAQTP---SGA
******: :*..* *** *:*: *:: * : * .: * :* :
MSMEG_5507|M.smegmatis_MC2_155 QVDLLALSLQGLYQAFFQGYGGYANPGTEEVTTYERNPDGTLVLDANGDP
TH_4419|M.thermoresistible__bu GSGFPIAPSPAPVEDLATGLHLVEDVG---------GPDAPTLLLADPST
.: . . : : * : * .**.. :* *: ..
MSMEG_5507|M.smegmatis_MC2_155 IPITTEEPVFGDCTATGAVCLSGFPGAAYYLSDELLPLGDVDNIFFESGI
TH_4419|M.thermoresistible__bu LQTLLDAFFIGYPTSAGP---DGFTGVAYFAVDQIFPDSPIVDAFFEGGF
: : .:* *::*. .**.*.**: *:::* . : : ***.*:
MSMEG_5507|M.smegmatis_MC2_155 EPFFQIAAQTIAAEVDALDPTGRLTLERRVADFFEGGATQLVGNTLLDNL
TH_4419|M.thermoresistible__bu TEVARVVLVGLAGGEGTP-------LGGAINDFFEGGVTQLVGTALVDAL
. ::. :*. .: * : ******.*****.:*:* *
MSMEG_5507|M.smegmatis_MC2_155 SEGTVAYGLTNSFFFGYGEN-------AGVVAALTYVVDAIAQGTADPVP
TH_4419|M.thermoresistible__bu PDDSYAHGVTDAFFFGYSNDPDDDEPPNGIVGATYYTLTTLISGTPPPVP
.:.: *:*:*::*****.:: *:*.* *.: :: .**. ***
MSMEG_5507|M.smegmatis_MC2_155 NPINPLGPGVSSATVETQLVSKQAIEQKVVEGEDPASTNILTRSRFSVPV
TH_4419|M.thermoresistible__bu DELA--EPSTFGAVPFAATGRIPAVDQNPGPRVDGDTIGKTPVPAVPPTA
: : *.. .*. : *::*: * : . . . .. ..
MSMEG_5507|M.smegmatis_MC2_155 SGEALGFKSDANTTPVLEKDGLGEETTSDGVAPAVNVTSPQSAALGQAPE
TH_4419|M.thermoresistible__bu AVNPGGVFENPVESPPASGRSWRVPTPPQ---PDPEPAAPPTDAAVAAEE
: :. *. .:. :* . . *..: * : ::* : * * *
MSMEG_5507|M.smegmatis_MC2_155 TPATAPEEPSAPAAEVKEETESTGATANTKDGNKFTPGVILSPGGRKSSS
TH_4419|M.thermoresistible__bu TPEPASTGRKPAAVDRTKRNQKP--AVDMTTGNKTTVEPLLPFGKRPGGD
** .*. ...*.: .:..:.. :.: . *** * :*. * * ...
MSMEG_5507|M.smegmatis_MC2_155 ANGWEKLEQDVKKGINTLGKIFNKGKTSKPATSSTTDNAGEGGEGGGDE
TH_4419|M.thermoresistible__bu PG--NRIRDTWKR----------ITDPSTPARSPESESTDEG-------
.. :::.: *: ..*.** *. ::.:.**