For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GDRIPARLAEHPTVRAVRSRGAAGPRVPIDADWLRALCLEAGVDDVGFVSVDDPAVESERGHADAALPGV RSYISLVVRMNRDNVRSTARSVANQEFHRSGEALNEAAHRITRSLEDAGYRAVNPSATFPMEMDRYPGRI WVIAHKPVAVAAGMGVMGIHRNVIHPKYGNFILLGTILVGAPITSYGEPLDYSPCLECKLCVAVCPVGAI GKNGEFDFRACSVHNYREFMGGFTDWVQTVADSADADDFRSRVSDSENASMWQSLSFKPNYKAAYCLAVC PAGEDVIEPYLDDRKAFMDLVLKPLQDKKETLYVLPNSAAKEYAEKRYPHKTVKVVDGGLPRGQ*
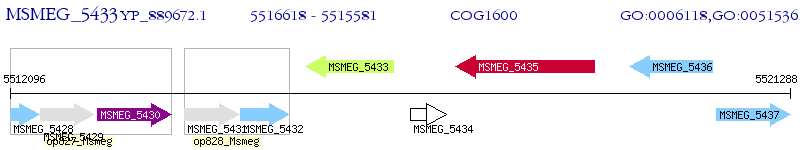
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5433 | - | - | 100% (345) | 4Fe-4S ferredoxin iron-sulfur binding domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4789 | - | 1e-164 | 80.65% (341) | 4Fe-4S ferredoxin iron-sulfur binding domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5433|M.smegmatis_MC2_155 MGDRIPARLAEHPTVRAVRSRGAAGPRVPIDADWLRALCLEAGVDDVGFV
Mvan_4789|M.vanbaalenii_PYR-1 MLEKLPVRLAEHPTVRAVRSRPAQAPGV-IDADWLRKVCLDAGADDVGFA
* :::*.************** * .* * ******* :**:**.*****.
MSMEG_5433|M.smegmatis_MC2_155 SVDDPAVESERGHADAALPGVRSYISLVVRMNRDNVRSTARSVANQEFHR
Mvan_4789|M.vanbaalenii_PYR-1 SVGDPALSSELAHVEAALPGAVSYISLVVKMNRDNVRSSARSVANQEFHR
**.***:.** .*.:*****. *******:********:***********
MSMEG_5433|M.smegmatis_MC2_155 SGEALNEAAHRITRSLEDAGYRAVNPSATFPMEMDRYPGRIWVIAHKPVA
Mvan_4789|M.vanbaalenii_PYR-1 SGEVMNEAAHRIARVLQDAGHRVVNPSATFPMEMDRFPGRIWVVAHKPVA
***.:*******:* *:***:*.*************:******:******
MSMEG_5433|M.smegmatis_MC2_155 VAAGMGVMGIHRNVIHPKYGNFILLGTILVGAPITSYGEPLDYSPCLECK
Mvan_4789|M.vanbaalenii_PYR-1 VAAGLGAMGIHRNVIHPRFGNFILLGTVLVASEISSYGTPLDYSPCLECK
****:*.**********::********:**.: *:*** ***********
MSMEG_5433|M.smegmatis_MC2_155 LCVAVCPVGAIGKNGEFDFRACSVHNYREFMGGFTDWVQTVADSADADDF
Mvan_4789|M.vanbaalenii_PYR-1 LCVAACPVGAIKKNGDFDFVACSVHNYREFMGGFTDWAQTIADSADAAEF
****.****** ***:*** *****************.**:****** :*
MSMEG_5433|M.smegmatis_MC2_155 RSRVSDSENASMWQSLSFKPNYKAAYCLAVCPAGEDVIEPYLDDRKAFMD
Mvan_4789|M.vanbaalenii_PYR-1 RSRVSDSENASMWQSLSFKANYKAAYCLAVCPAGEDVIEPYLDDRKGFMD
*******************.**************************.***
MSMEG_5433|M.smegmatis_MC2_155 LVLKPLQDKKETLYVLPNSAAKEYAEKRYPHKTVKVVDGGLPRGQ
Mvan_4789|M.vanbaalenii_PYR-1 LVLKPLQDKVETLYVLPNSDAEAHAVRRYPHKPVKRVDSGI-RGV
********* ********* *: :* :*****.** **.*: **