For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAEPLLVVGLGNPGPTYAKTRHNLGFMVADVLAGRIGSAFKVHKKSGAEVVTGRLAGTSVVLAKPRCYMN ESGRQVGPLAKFYSVPPQQIVVIHDELDIDFGRIRLKLGGGEGGHNGLRSVASALGTKNFHRVRIGVGRP PGRKDPAAFVLENFTAAERAEVPTIVEQAADATELLIAQGLEPAQNTVHAW
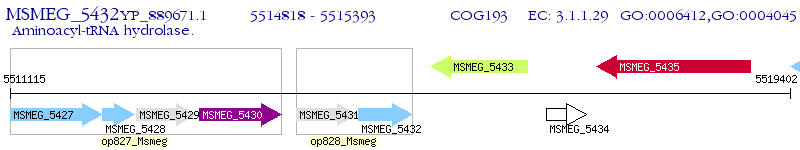
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5432 | pth | - | 100% (191) | peptidyl-tRNA hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1042c | pth | 1e-91 | 82.20% (191) | peptidyl-tRNA hydrolase |
| M. gilvum PYR-GCK | Mflv_1938 | - | 5e-84 | 77.49% (191) | peptidyl-tRNA hydrolase |
| M. tuberculosis H37Rv | Rv1014c | pth | 1e-91 | 82.20% (191) | peptidyl-tRNA hydrolase |
| M. leprae Br4923 | MLBr_00244 | pth | 8e-82 | 75.00% (188) | peptidyl-tRNA hydrolase |
| M. abscessus ATCC 19977 | MAB_1142c | - | 2e-76 | 71.43% (189) | peptidyl-tRNA hydrolase (PTH) |
| M. marinum M | MMAR_4473 | pth | 2e-91 | 83.25% (191) | peptidyl-tRNA hydrolase Pth |
| M. avium 104 | MAV_1151 | pth | 7e-92 | 82.20% (191) | peptidyl-tRNA hydrolase |
| M. thermoresistible (build 8) | TH_0906 | pth | 1e-22 | 79.66% (59) | peptidyl-tRNA hydrolase |
| M. ulcerans Agy99 | MUL_4644 | pth | 3e-92 | 83.77% (191) | peptidyl-tRNA hydrolase |
| M. vanbaalenii PYR-1 | Mvan_4788 | - | 7e-87 | 79.58% (191) | peptidyl-tRNA hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1938|M.gilvum_PYR-GCK ---MAEP--------VLVVGLGNPGPQYATTRHNIGFMVADVLADRMGET
Mvan_4788|M.vanbaalenii_PYR-1 ---MAEP--------VLVVGLGNPGPQYATTRHNAGFMVVDILADRMGEK
Mb1042c|M.bovis_AF2122/97 ---MAEP--------LLVVGLGNPGANYARTRHNLGFVVADLLAARLGAK
Rv1014c|M.tuberculosis_H37Rv ---MAEP--------LLVVGLGNPGANYARTRHNLGFVVADLLAARLGAK
MAV_1151|M.avium_104 ---MAEP--------LLVVGLGNPGDNYARTRHNVGFMVADLLAARMGSK
MMAR_4473|M.marinum_M ---MAEP--------LLVVGLGNPGENYARTRHNLGFMVADLLAARLGAK
MUL_4644|M.ulcerans_Agy99 ---MAEP--------LLVVGLGNPGENYARTRHNLGFMVADLLAARLGAK
MLBr_00244|M.leprae_Br4923 ---MAEPTLASGWYPRLVVGLGNPGKNYGRTRHNVGFMVANLLAVRLGSK
MSMEG_5432|M.smegmatis_MC2_155 ---MAEP--------LLVVGLGNPGPTYAKTRHNLGFMVADVLAGRIGSA
MAB_1142c|M.abscessus_ATCC_199 MPVLSDD-------AVLVVGLGNPGPNYAKTRHNLGFMVADLLAAKDGAT
TH_0906|M.thermoresistible__bu ---MSEPQPAAG--PRLIVGLGNPGPRYAGTRHNLGFMVADILAGRMGEK
::: *:******* *. **** **:*.::** : *
Mflv_1938|M.gilvum_PYR-GCK FKVHKKSGAEVTTGRLAGRPVVLAKPRVYMNESGRQVGPLAKFYSIAPTD
Mvan_4788|M.vanbaalenii_PYR-1 FKVHKKSGAEVATGRLAGRPVVLAKPRVYMNESGRQVGPLAKFYSVAPAD
Mb1042c|M.bovis_AF2122/97 FKAHKRSGAEVATGRSAGRSLVLAKPRCYMNESGRQIGPLAKFYSVAPAN
Rv1014c|M.tuberculosis_H37Rv FKAHKRSGAEVATGRSAGRSLVLAKPRCYMNESGRQIGPLAKFYSVAPAN
MAV_1151|M.avium_104 FKAHKRSGAEIVSGRLAGRSVVVAKPRCYMNESGRQVGPLAKFYSVPPGD
MMAR_4473|M.marinum_M FKVHKRSGAEVVTGRLAQRSVVLAKPRCYMNESGRQVAPLAKFYSVPPAD
MUL_4644|M.ulcerans_Agy99 FKVHKRSGAEVVTGRLAQRSVVLAKPRCYMNESGRQVAPLAKFYSVPPAD
MLBr_00244|M.leprae_Br4923 FEVHKRSGADVVNGRLAGCSMLVAKPRNYMNESGQQVGLLAKLYSVTPAD
MSMEG_5432|M.smegmatis_MC2_155 FKVHKKSGAEVVTGRLAGTSVVLAKPRCYMNESGRQVGPLAKFYSVPPQQ
MAB_1142c|M.abscessus_ATCC_199 FKPHKKSGAEATTIRLGGRAVNLAKPRTFMNTSGPQVAALAKFFSVPPAN
TH_0906|M.thermoresistible__bu FKVHKKSGAEVITGRLAGRPVXX---------------------------
*: **:***: . * . .:
Mflv_1938|M.gilvum_PYR-GCK VVIIHDELDIDFGRIRLKAGGGVAGHNGLRSVGSALSTNDFQRVRVGIGR
Mvan_4788|M.vanbaalenii_PYR-1 VVIVHDELDIDFGRIRLKAGGGVAGHNGLRSVASALGGNDFQRVRVGIGR
Mb1042c|M.bovis_AF2122/97 IIVIHDDLDLEFGRIRLKIGGGEGGHNGLRSVVAALGTKDFQRVRIGIGR
Rv1014c|M.tuberculosis_H37Rv IIVIHDDLDLEFGRIRLKIGGGEGGHNGLRSVVAALGTKDFQRVRIGIGR
MAV_1151|M.avium_104 IIVIHDDLDLDFGRIRLKIGGGEGGHNGLRSVSNALGSKDFQRVRIGIGR
MMAR_4473|M.marinum_M LIVIHDELDLDFGRIRLKFGGGEGGHNGLRSVAAALGTKDFQRVRIGIGR
MUL_4644|M.ulcerans_Agy99 LIVIHDELDLDFGRIRLKFGGGEGGHNGLRSVAAALGTKNFQRVRIGIGR
MLBr_00244|M.leprae_Br4923 IIIVHDDLDLDFGRIRLKLGGGEGGHNGLRSVAAALGTKDFQRVRIGIGR
MSMEG_5432|M.smegmatis_MC2_155 IVVIHDELDIDFGRIRLKLGGGEGGHNGLRSVASALGTKNFHRVRIGVGR
MAB_1142c|M.abscessus_ATCC_199 VIVMHDELDLDFGTVRLKLGGGEGGHNGLRSVSQSLGTKDYLRVRLGVGR
TH_0906|M.thermoresistible__bu -------------------GGSPEQHPDL----------DLLRQRQ--AR
**. * .* : * * .*
Mflv_1938|M.gilvum_PYR-GCK PPGQKSGASFVLEPFNSRERPELGTIIEQAADATELLIELGIEPAQNTVH
Mvan_4788|M.vanbaalenii_PYR-1 PPGHKSGASFVLENFNSVERKEVPTILEQAADATELLVAQGLEPAQNTVH
Mb1042c|M.bovis_AF2122/97 PPGRKDPAAFVLENFTPAERAEVPTICEQAADATELLIEQGMEPAQNRVH
Rv1014c|M.tuberculosis_H37Rv PPGRKDPAAFVLENFTPAERAEVPTICEQAADATELLIEQGMEPAQNRVH
MAV_1151|M.avium_104 PPGRKDPAAFVLENFTSTERADVPTICEQAADATELLIELGLEPAQNRVH
MMAR_4473|M.marinum_M PPGRKDPATFVLENFSSPERPEVPTICEQAADATELLVEIGLEPAQNRVH
MUL_4644|M.ulcerans_Agy99 PPGRKDPATFVLENFSSPERPEVPTICEQAADATELLVEVGLEPAQNRVH
MLBr_00244|M.leprae_Br4923 PTGRKDPASFVLENFTTAERMQVPTICKRAADATELLVGLGLEPAQNHVH
MSMEG_5432|M.smegmatis_MC2_155 PPGRKDPAAFVLENFTAAERAEVPTIVEQAADATELLIAQGLEPAQNTVH
MAB_1142c|M.abscessus_ATCC_199 PPGRKDPAAFVLENFAKAETDQVPLLCEQGADAAELLIRLGLEAAQNQVH
TH_0906|M.thermoresistible__bu --------------------------------------------------
Mflv_1938|M.gilvum_PYR-GCK AWS
Mvan_4788|M.vanbaalenii_PYR-1 AWG
Mb1042c|M.bovis_AF2122/97 AW-
Rv1014c|M.tuberculosis_H37Rv AW-
MAV_1151|M.avium_104 AW-
MMAR_4473|M.marinum_M AW-
MUL_4644|M.ulcerans_Agy99 AW-
MLBr_00244|M.leprae_Br4923 AW-
MSMEG_5432|M.smegmatis_MC2_155 AW-
MAB_1142c|M.abscessus_ATCC_199 AWG
TH_0906|M.thermoresistible__bu ---