For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNTERSLWNWDYAHEVLPGLVAAFFKFTLGITAASSLVAIVLGLVLVLGRRSGARWISWPSYAVVEFIRS TPIPIQLFFVYFGLPVVGIRPSALVTGIAVLGVHYACYMSEVYRAGIDAIPQGQWEACTALSLSPYRTWR AVVLPQVVRRILPSTGNQVIALFKETPFLIVIGVLEMVTVANQFGGQNYRYIEPITIAGLIFLAASYPTS VLMRKLEIRVAR
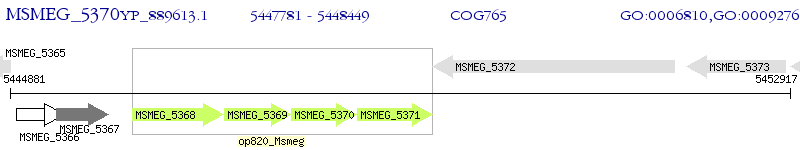
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5370 | ehuD | - | 100% (222) | ectoine/hydroxyectoine ABC transporter, permease protein EhuD |
| M. smegmatis MC2 155 | MSMEG_3549 | ehuD | 3e-47 | 43.72% (215) | ectoine/hydroxyectoine ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_5318 | - | 1e-27 | 32.83% (198) | glutamine ABC transporter, permease/substrate-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3164 | - | 1e-29 | 32.84% (204) | polar amino acid ABC transporter, inner membrane subunit |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0277c | - | 1e-24 | 28.57% (203) | amino acid ABC transporter permease |
| M. marinum M | MMAR_0061 | glnQ | 3e-27 | 30.36% (224) | ABC transporter permease protein GlnQ |
| M. avium 104 | MAV_0059 | - | 2e-28 | 31.70% (224) | amino acid ABC transporter, permease protein, |
| M. thermoresistible (build 8) | TH_0515 | - | 3e-26 | 31.86% (226) | amino acid ABC transporter, permease protein, 3-TM region, |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_6047 | - | 4e-29 | 32.14% (224) | polar amino acid ABC transporter, inner membrane subunit |
CLUSTAL 2.0.9 multiple sequence alignment
TH_0515|M.thermoresistible__bu -----VTRAAVV-AVIALLVGSLTAGGT--VLAAPAAATVDQCAPPGVQS
Mvan_6047|M.vanbaalenii_PYR-1 MSCMRVLRPSVKSLVLALIIGMVAAMG----LAAPAGAQVDQCTPPGIES
MMAR_0061|M.marinum_M --------------MLAVSATILIVLG--LALAAPALADHNQCAPAGVDS
MAV_0059|M.avium_104 ----MGRRGTRRGKLLALTATVLMVCGSSALLAPPAAADRNQCAPAGPKS
MSMEG_5370|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3164|M.gilvum_PYR-GCK ---------------------------------MGAAEGSSGCRPGPPGN
MAB_0277c|M.abscessus_ATCC_199 --------------------------------------------------
TH_0515|M.thermoresistible__bu ASPLPTNLAAAEAGP-GVDRYTTDGVVPLDNIDPSALRLRVPGVLTVGTL
Mvan_6047|M.vanbaalenii_PYR-1 ASALPTNLAAAAAGP-AADKYTTATVKPLASVDLNALGLTQPGTLTVGTL
MMAR_0061|M.marinum_M AMVLPADLSSATTAGSDEDVQTTAGVVPLSSVNIGDLGLGTPGVLTAGTL
MAV_0059|M.avium_104 AAVLPENLTRGGSMS-QPDEHTTPTVEPLSSVRIDTLGLGTPGVLTVGTL
MSMEG_5370|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3164|M.gilvum_PYR-GCK RRRRLQILIAGLLIG-----------LFGAVLALPGTAGAEGETYTVATD
MAB_0277c|M.abscessus_ATCC_199 -------------------MFRAIVLALLALVAVAGCAAPTQEPLRVGTE
TH_0515|M.thermoresistible__bu SDAPPSICIN-SLGEFTGFDNELLRAIAAKLGLQISFVGTEFSALLAQVA
Mvan_6047|M.vanbaalenii_PYR-1 SDAPPSICID-SAGQYTGFDNELLKAVAEKLGLRVNFVGTEFSGLLAQVA
MMAR_0061|M.marinum_M SDAPPNTCIN-STGQFTGFDNELLRAIAAKLGLQVRFVGTDFSGLLAQVA
MAV_0059|M.avium_104 SGAPPNVCIT-PTGQYSGFDNQLLRAIAGKLGLQVRFVGTDFAGLLAQVA
MSMEG_5370|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3164|M.gilvum_PYR-GCK TTFAPFEFQD-ASGEFVGIDMDLIRAIAEDQGFTVDIKPLGFDAALQAVQ
MAB_0277c|M.abscessus_ATCC_199 GVYAPFSFHDGHTGELTGYDVDVARAVGEKLGRPVEFVEIPWDAIFAGLD
TH_0515|M.thermoresistible__bu SGRFDVGSSSITTTEQRRRTVEFTNGYDFGYFSLVVPSGSAITGFGDLVP
Mvan_6047|M.vanbaalenii_PYR-1 ARRFDVGSSSITTTDARRRTVGFTNGYDFGYFSLVVPKGSAITGFEKLAP
MMAR_0061|M.marinum_M ARRFDVGSASVKLTEARRRTVAFTNGYDFGYYSLVVPPGSAITKFSDLAP
MAV_0059|M.avium_104 SRRFDVGSSSIKATDERRQTVAFTNGYDFGYYALVVPPGSAIKEFTDLAA
MSMEG_5370|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3164|M.gilvum_PYR-GCK ANQVDGVIAGMSITEERKKVFDFSEPYFESGIQMAVLEDNEDITSYEDLR
MAB_0277c|M.abscessus_ATCC_199 AERFDVVANQVTITPARKAKYDISTPYAVGEGVIVTRADDNSIHSLSDVR
TH_0515|M.thermoresistible__bu GQRIGVVQGTVQESYV---VDTLGLRPVIFPDYNTVYASLKTRQIDAWVA
Mvan_6047|M.vanbaalenii_PYR-1 GQRIGVVQGTVQEAYV---IDTLGLQPVKFPDYNTVYASLKTGQIDAWVA
MMAR_0061|M.marinum_M GQRIGVVQGTVEESYV---VNSLHLQPVKYPDFATVYGNLKTHQIDAWVA
MAV_0059|M.avium_104 GQRIGVVQGTVEESYV---VDTLHLQPVKYPDFATVYASLKTRQIDAWVA
MSMEG_5370|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3164|M.gilvum_PYR-GCK GQRVSVKNGTQGSDFANSIKDQYGFEVVSFADSSTMFDEVKTGNSVAVFE
MAB_0277c|M.abscessus_ATCC_199 GKVAAENATSNWSQIAR----DAGARVEAVEGFTQSITLLSQGRVDVVIN
TH_0515|M.thermoresistible__bu PSQQAEGTVRPGDPAEIIENTFSLDNFIAYAVAPGNQPLIDALNAGLDAV
Mvan_6047|M.vanbaalenii_PYR-1 PSQQAQGTVQPGDPADIIENTFSLDNFIAWAVAGDNQPLIDALNSGLDAV
MMAR_0061|M.marinum_M PGNQAEAAIRPGDPAVLVANTFGAGDFVAYVVARDNRPLLDALNSGLDAV
MAV_0059|M.avium_104 PAGQAATTIQPGDPAVVVANTISPGNFVAYAVAKDNKPLVDALNSGLDAV
MSMEG_5370|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3164|M.gilvum_PYR-GCK DYPVLLYGIAQGNGFKTVTPKEDPTGYGFAVNKGQNAELLQKFNDGLENL
MAB_0277c|M.abscessus_ATCC_199 DSIAVYAYLASTGDPAVKIAGATGQRSEQGFAARKNSGLLPDLDKALDQL
TH_0515|M.thermoresistible__bu IADGTWARLYSEWVPRALPPGWKPGSKAAPPPQLPDFDAIAAERASQQPA
Mvan_6047|M.vanbaalenii_PYR-1 IADGTWSRLYTDWVPRAVPPGWKPGSKAAPAPELPDFAAIAAQNQSAQGP
MMAR_0061|M.marinum_M IADGTWSQLYSKWVPRTLPPGWKPGSRAVAAPTLPDFAAIAAAHR--HKP
MAV_0059|M.avium_104 IADGTWSNLYSQWVPRTLPPGWKPGSQAVAPPKLPDFAAIAASQH--HKA
MSMEG_5370|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3164|M.gilvum_PYR-GCK KAQGRYDEILQTYLG-----------------------------------
MAB_0277c|M.abscessus_ATCC_199 AADGTLTRISQKYLKTDAS-------------------------------
TH_0515|M.thermoresistible__bu AGPGVRKSTLAQLADSFLDWELYRQAIPDLLTTGLPNTLLLTVGAGVIGL
Mvan_6047|M.vanbaalenii_PYR-1 TAPVQPKSTLSQLAASFLDWDLYKQAIPDLFKTGLPNTLILTVAASIIGL
MMAR_0061|M.marinum_M VGPAAPKSALTQLRDSFFDWDMYRQAIPTLFKTGLPNTLILTLSASIIGL
MAV_0059|M.avium_104 VGPAAPKSTLAQLRDSFFDWDMYRQAIPTLLRTGLPNTLILTFSASVIGL
MSMEG_5370|M.smegmatis_MC2_155 ----------MNTERSLWNWDYAHEVLPGLVAAFFKFTLGITAASSLVAI
Mflv_3164|M.gilvum_PYR-GCK ----------EEAAESDDSLFGLIKSTSPMLLAGLKMTIILTVVSIFFAL
MAB_0277c|M.abscessus_ATCC_199 -GTTKPQAGKGDSGSTRSDTQLVLDNLWPMARAMITVTLPLTAISFVIGL
: : . . : : : *: :* : ...:
TH_0515|M.thermoresistible__bu VLGMVLAIAGISRSRLLRWPARVYTDIFRGLPEVVIILLIGLGLGPVVGG
Mvan_6047|M.vanbaalenii_PYR-1 ILGMALAIAGISKSRWLRWPARIYTDIFRGLPEVVIILLIGLGVGPVVGH
MMAR_0061|M.marinum_M ALGMALAIAGISHQRWLRWPARVYTDIFRGLPEVVIILIIGLGVGPIVGG
MAV_0059|M.avium_104 VLGMVLAVAGISHARWLRWPARVYTDVFRGLPEVVIILLIGLGIGPIVGG
MSMEG_5370|M.smegmatis_MC2_155 VLGLVLVLGRRSGARWISWPSYAVVEFIRSTPIPIQLFFVYFGL-PVVG-
Mflv_3164|M.gilvum_PYR-GCK ILGVIFGLARVSRSIWLRAIGTTFVDIFRGTPLLVQAFFIYFGIPATMG-
MAB_0277c|M.abscessus_ATCC_199 VIALGVALARMSGHRVVTGLARIYISVIRGTPLLLQLFLIFFAL-PEFG-
:.: . :. * : . ..:*. * : ::: :.: . .*
TH_0515|M.thermoresistible__bu LTGYNPFPLGILALGLMAAAYVGEILRSGIQSVEPGQLEAARALGFSYPA
Mvan_6047|M.vanbaalenii_PYR-1 LTGNNPFPLGIAALGLMAAAYVGEIFRSGIQSVDPGQLEASRALGFSYPT
MMAR_0061|M.marinum_M LTNNNPYPLGIAALGLMAGAYVGEILRSGIQSVDPGQLEASRALGFSYSA
MAV_0059|M.avium_104 LTNNNPYPLGIAALGMMAAAYIGEILRSGIQSVDPGQLEASRALGFSYAT
MSMEG_5370|M.smegmatis_MC2_155 -IRPSALVTGIAVLGVHYACYMSEVYRAGIDAIPQGQWEACTALSLSPYR
Mflv_3164|M.gilvum_PYR-GCK -FQMSAVTAGIITLSLNAGAYMTEIVRGGILSVDKGQMEAARSLGIGYLP
MAB_0277c|M.abscessus_ATCC_199 -VKISPFPAAVIAFSLNVGGYAAEIIRSSILSIPRGQWEAASSLGMSYST
.. .: .:.: . * *: *..* :: ** **. :*.:.
TH_0515|M.thermoresistible__bu AMRLVVVPQGVRRVLPALMNQFISLLKASSLVYFLGLVADQRELFQVGRD
Mvan_6047|M.vanbaalenii_PYR-1 SMRLVVVPQGVRRVLPALMNQFISLLKASSLVYFLGLIANQRELFQVGRD
MMAR_0061|M.marinum_M AMRLVVVPQGVRRVLPALVNQFIALLKASALVYFLGLIAQQRELFQVGRD
MAV_0059|M.avium_104 AMRLVVVPQGVRRVLPALVNQFIALLKASALVYFLGLVADQRELFQVGRD
MSMEG_5370|M.smegmatis_MC2_155 TWRAVVLPQVVRRILPSTGNQVIALFKETPFLIVIGVLEMVTVANQFG--
Mflv_3164|M.gilvum_PYR-GCK TMRKVILPQAIRTMIPSYINQFVITLKDTSILSVLGIAELTQ---SGRII
MAB_0277c|M.abscessus_ATCC_199 TLWRIVIPQASRVAVPPLSNTLISLVKDTSLASAILVTDVMR---TAQVA
: :::** * :*. * .: .* :.: : :
TH_0515|M.thermoresistible__bu LNAQTGSLSPLVAAGLFYLALTIPLTHLVNYVDNRLRRGRK-PAAEDPLE
Mvan_6047|M.vanbaalenii_PYR-1 LNAQTGNLSPLVAAGLFYLALTIPLTHLVNFIDTRLRRGRK-PDEKDPLV
MMAR_0061|M.marinum_M LNAQTGNLSPLVAAGIFYLMLTVPLTHLVNYIDARLRRGRRRLDPEDPPG
MAV_0059|M.avium_104 LNAETGNLSPLVAAGACYLILTVPLTHLVNMIDARLRRGRAAVDPEEPAE
MSMEG_5370|M.smegmatis_MC2_155 -GQNYRYIEPITIAGLIFLAASYPTSVLMRKLEIRVAR------------
Mflv_3164|M.gilvum_PYR-GCK IAGNYKAFEMWLIVGVIYFIVIMALTKLSDRLERRLVK------------
MAB_0277c|M.abscessus_ATCC_199 AAPTFQFFTLYVTAGVYYWIVCMAMSAVQDRLEQRLEKSVAR--------
: .* : . : : :: *: :
TH_0515|M.thermoresistible__bu PAT---AQEMI
Mvan_6047|M.vanbaalenii_PYR-1 LAT---SQEMN
MMAR_0061|M.marinum_M VVSSTIGEEMT
MAV_0059|M.avium_104 MLT--FGQEIT
MSMEG_5370|M.smegmatis_MC2_155 -----------
Mflv_3164|M.gilvum_PYR-GCK -----------
MAB_0277c|M.abscessus_ATCC_199 -----------