For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTETQLDRTGAATIVTEHPDQFRIAVDGKTVGVADYHDRDGRRIFPHTEVLPQYRGRGLATIMIAEALR STRAAGLRIVPTCWMVAEFIDKHPEYADITDRR
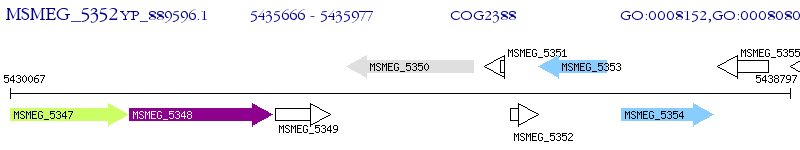
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5352 | - | - | 100% (103) | hypothetical protein MSMEG_5352 |
| M. smegmatis MC2 155 | MSMEG_0356 | - | 8e-13 | 37.97% (79) | acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2008 | - | 2e-43 | 75.49% (102) | hypothetical protein Mflv_2008 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3942 | - | 3e-13 | 42.22% (90) | hypothetical protein MAB_3942 |
| M. marinum M | MMAR_0039 | - | 4e-28 | 59.57% (94) | hypothetical protein MMAR_0039 |
| M. avium 104 | MAV_0037 | - | 7e-25 | 53.19% (94) | hypothetical protein MAV_0037 |
| M. thermoresistible (build 8) | TH_0230 | - | 9e-26 | 56.38% (94) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0038 | - | 7e-26 | 55.32% (94) | hypothetical protein MUL_0038 |
| M. vanbaalenii PYR-1 | Mvan_4721 | - | 4e-41 | 72.55% (102) | hypothetical protein Mvan_4721 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2008|M.gilvum_PYR-GCK MRDYREVMSSEMPPDRTGAATTVTREPDRFVIEVEGRAVGLADYHDTDGR
Mvan_4721|M.vanbaalenii_PYR-1 -------MTSETPPDRTGAHTAVTRQSGRFVIEVEGRPVGLADYHDHDGR
MSMEG_5352|M.smegmatis_MC2_155 -------MSTETQLDRTGAATIVTEHPDQFRIAVDGKTVGVADYHDRDGR
MMAR_0039|M.marinum_M -------MT----ADRTGAQTTITAEAGKYTIAVEGKTVGLATYIDRGNQ
MUL_0038|M.ulcerans_Agy99 -------MT----ADKTGAQTTITAEAGKYTIAIEGQTVELATYIDRGDQ
MAV_0037|M.avium_104 -------MT----TDKTGAQTTVSLQDGKYTIAVEGKPVGLADFKDRGDQ
TH_0230|M.thermoresistible__bu -------MT----TDKTGAPTTVAPGDGRFTIAVKNKSVGSADFIDHGDQ
MAB_3942|M.abscessus_ATCC_1997 -------------MDESDVTVADAPDNHRFEITADGELAGFTEYLDTSLD
*.:.. . : :: * ... . : : * .
Mflv_2008|M.gilvum_PYR-GCK ----RVFPHTEVLPQFQGRGLATILVAEALRVTKSEGLRIVPTCWMVAEY
Mvan_4721|M.vanbaalenii_PYR-1 ----RVFPHTEVLPQFQGRGLATILVAEALRVTQAEGLRIVPTCWMVAEY
MSMEG_5352|M.smegmatis_MC2_155 ----RIFPHTEVLPQYRGRGLATIMIAEALRSTRAAGLRIVPTCWMVAEF
MMAR_0039|M.marinum_M ----RVFDHTEVDPAFGGRGLATILVEEALQDARAAGKRIVPVCSMVVTV
MUL_0038|M.ulcerans_Agy99 ----RVFDHTEVDPAFGGRGLATILVEEALQDARAAGKRIVPVCSMVVTV
MAV_0037|M.avium_104 ----RVFYHTEIDPAYGGRGLATILVEEALNEARNEGKRIVPVCSMIGTV
TH_0230|M.thermoresistible__bu ----RVFTHTTVDSAFEGRGLATILIGEALRQTRDAGLRIVPVCQMVQNY
MAB_3942|M.abscessus_ATCC_1997 GTAVRIFFHTEIDEKFGGRGLAGTLVQSALTTSRGTGRKIVPVCPYVKKF
*:* ** : : ***** :: .** :: * :***.* :
Mflv_2008|M.gilvum_PYR-GCK IDKHPEYADITDRS-------------
Mvan_4721|M.vanbaalenii_PYR-1 IDRHPEYADITDRT-------------
MSMEG_5352|M.smegmatis_MC2_155 IDKHPEYADITDRR-------------
MMAR_0039|M.marinum_M LDKHPEYDEITDPV-------------
MUL_0038|M.ulcerans_Agy99 LDKHPEYDAITDPV-------------
MAV_0037|M.avium_104 LKKHPEFDDITDPVTPEITQWVHS---
TH_0230|M.thermoresistible__bu VGKHPEYTDVTDAATEEHRRLADS---
MAB_3942|M.abscessus_ATCC_1997 VGKHHEFDDIVTPVTRDALAAVEARQG
: :* *: :.