For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ERLFSLVVGVLMAVAGAAAPPAQAQDFAITDEVPTLEELTSQLQLLVATQAPDHVKAAQLEGGSRALIVP KMVYRLGVLRAPRGYVEVTGPETHEEGRHTAVINGHRQGSPTIVVPAEWRFIDGRWKLASKSMCNGISAL GLPIPCNFQ*
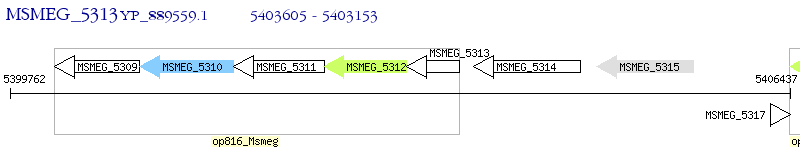
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5313 | - | - | 100% (150) | hypothetical protein MSMEG_5313 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0001 | - | 1e-51 | 68.61% (137) | hypothetical protein MSMEG_5313 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2046 | - | 6e-52 | 64.71% (153) | hypothetical protein Mvan_2046 |
CLUSTAL 2.0.9 multiple sequence alignment
TH_0001|M.thermoresistible__bu -------VIAALVAAVTTASLGFAPTASAQPGQPTG--SVEDFAITSEVP
Mvan_2046|M.vanbaalenii_PYR-1 MMRWCRGAAAALFAAATVP-LTLAPGAAADPEIHADPQGPQEFAITSEVP
MSMEG_5313|M.smegmatis_MC2_155 --------MERLFSLVVGVLMAVA-GAAAPPA------QAQDFAITDEVP
*.: .. : .* *:* * ::****.***
TH_0001|M.thermoresistible__bu TLEELDAQIRLLVGTAAPDWVKAAQLEGGDRAVIVPKMVYRIGFFRPPRG
Mvan_2046|M.vanbaalenii_PYR-1 TLAELDAQIRLLVATPAPDWVKAAQLEGGDRAVVVPKMIYRVGFFRSPKG
MSMEG_5313|M.smegmatis_MC2_155 TLEELTSQLQLLVATQAPDHVKAAQLEGGSRALIVPKMVYRLGVLRAPRG
** ** :*::***.* *** *********.**::****:**:*.:*.*:*
TH_0001|M.thermoresistible__bu SSRVTGPETHDGDRHTAVINASRQGAPTVRVEAEWRRIDGRWKLANKSLC
Mvan_2046|M.vanbaalenii_PYR-1 SSMVTGPETHEPDRHTAVINASRQGAPTVAVVAEWRRIDGRWKLASKSLC
MSMEG_5313|M.smegmatis_MC2_155 YVEVTGPETHEEGRHTAVINGHRQGSPTIVVPAEWRFIDGRWKLASKSMC
*******: .*******. ***:**: * **** ********.**:*
TH_0001|M.thermoresistible__bu NGIETMGLPIPCNFE
Mvan_2046|M.vanbaalenii_PYR-1 NGIKTIGLPIPCNFQ
MSMEG_5313|M.smegmatis_MC2_155 NGISALGLPIPCNFQ
***.::********: