For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NGVLTRSLRAEMVRTGGRGPLWLVLLPAAALIPVAITFLIACVAERFARIPGQQYVQQVPTTNAAYWVIN VTVVLVAVAAAYGQASESRLRATEYVWLALPRRPSVLAGKWVFYGLLGAVLSLVLVALVLTALPRVAPLV YGQVSVIDPVGLRLMWTVPVYAFFAAGLGVGVGVAVRTPAAAVGLLLFWVYVVEIAVGYLPSGYSLQRFM PFLNGIYATGQDIVLEPPWGRDAALVYVCGVFGLVLLCSMGFRRGAEK*
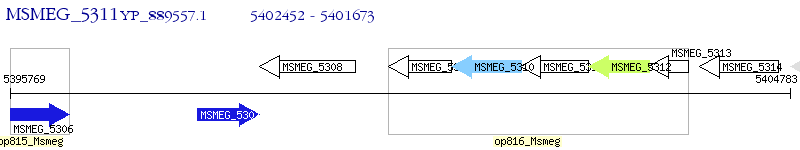
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5311 | - | - | 100% (259) | hypothetical protein MSMEG_5311 |
| M. smegmatis MC2 155 | MSMEG_0796 | - | 2e-14 | 29.11% (237) | ABC transporter integral membrane subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4300 | - | 3e-82 | 60.33% (242) | hypothetical protein Mflv_4300 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4214c | - | 8e-16 | 31.72% (186) | ABC transporter |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2659 | - | 6e-75 | 63.93% (219) | PUTATIVE hypothetical protein Mvan_2048 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2048 | - | 5e-83 | 60.66% (244) | hypothetical protein Mvan_2048 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4300|M.gilvum_PYR-GCK MSVALAVRGARAEMVRTGGRSRLWTVLIPAAVMLPAGITVAIAIAAETFA
Mvan_2048|M.vanbaalenii_PYR-1 -----MARGVRAELVRTSGRSRLWTVLVPAAVAVPAVITFAIALAAETFA
TH_2659|M.thermoresistible__bu -------------------------VIMPAAVGIPVVITLGIALIAEAFA
MSMEG_5311|M.smegmatis_MC2_155 -MNGVLTRSLRAEMVRTGGRGPLWLVLLPAAALIPVAITFLIACVAERFA
MAB_4214c|M.abscessus_ATCC_199 -----MGVVAAERIKVCTSRSAVWCSLL--ALALGVGLAGLQGGSASIYA
:: * : . :: . *. :*
Mflv_4300|M.gilvum_PYR-GCK RIPGQISVLQVPTSNAAYWVITVTTVVVALAASDGQACENRYHAKEYVRL
Mvan_2048|M.vanbaalenii_PYR-1 RIPGQISVLQVPTSNAAYWVITVTTVVVALGAADGQASENRYRAEEYVRL
TH_2659|M.thermoresistible__bu RIPGQISVLQVSTSNAAYWVITITTTLVAVAAADGQASESRHGTAEHVRL
MSMEG_5311|M.smegmatis_MC2_155 RIPGQQYVQQVPTTNAAYWVINVTVVLVAVAAAYGQASESRLRATEYVWL
MAB_4214c|M.abscessus_ATCC_199 PIS---------PGDAAGGAALVGVPLLMVLAAMTVTNENRTAMVRTTFQ
*. . :** . : . :: : *: : *.* . .
Mflv_4300|M.gilvum_PYR-GCK SAPRQWPVLTGRWLFYGALGAAAAAVALLTVLVLLPVVSPLVYGSVSVTD
Mvan_2048|M.vanbaalenii_PYR-1 AARRQWPVLVGRWLFHGALGAVVAVAALLVVLVALPVLSPHVYGSVSAAD
TH_2659|M.thermoresistible__bu AMPSRWSMLTARWVFYGGLGAVIAALTLIGVLSALPAVAPTVYGTVSWTD
MSMEG_5311|M.smegmatis_MC2_155 ALPRRPSVLAGKWVFYGLLGAVLSLVLVALVLTALPRVAPLVYGQVSVID
MAB_4214c|M.abscessus_ATCC_199 ATPNRLTVIGAKALVAGGWVAVVTAVTALTSMALVRAMAGPVAGANLALD
: : .:: .: :. * *. : : : :: * * *
Mflv_4300|M.gilvum_PYR-GCK PVALR-LLWTVPLLSFFAAGAGIGIGSLIRSPLGAAAVILLWVYAAESAA
Mvan_2048|M.vanbaalenii_PYR-1 PVALR-LLWTVPLLSFFAAGAGVGVGALIRSPLGAAAVILVWVYAFESAA
TH_2659|M.thermoresistible__bu PVGLR-LLWTVPLLAVFAAGAGVGVGALVRSPLAAVSAILLWAFVCETAA
MSMEG_5311|M.smegmatis_MC2_155 PVGLR-LMWTVPVYAFFAAGLGVGVGVAVRTPAAAVGLLLFWVYVVEIAV
MAB_4214c|M.abscessus_ATCC_199 STGVWNTVGAVTVYAFLCAVLAVAVGVLLKHTAGAVAVLLLWPTVLELML
...: : :*.: :.:.* .:.:* :: . .*.. :*.* . *
Mflv_4300|M.gilvum_PYR-GCK GYLP-SGAYLQRFMPVLNGVYATGQDAVLIP----PWSQNVALLYVCSLF
Mvan_2048|M.vanbaalenii_PYR-1 GYLP-SGAFLQRFMPVLNGVYATGQDAVLTP----PWGQNAALLYVCSLF
TH_2659|M.thermoresistible__bu GYLP-SGATLQRFMPMLNAVYATGQDTVLTP----PWGQNPALLYTCAVF
MSMEG_5311|M.smegmatis_MC2_155 GYLP-SGYSLQRFMPFLNGIYATGQDIVLEP----PWGRDAALVYVCGVF
MAB_4214c|M.abscessus_ATCC_199 GWLTDAGKALQPFLPFANGIRFTGVPWYTSDGVRFVWDATGSLIYFATVV
*:*. :* ** *:*. *.: ** *. :*:* . :.
Mflv_4300|M.gilvum_PYR-GCK TAIFVIAAVERTVRHG
Mvan_2048|M.vanbaalenii_PYR-1 TAIFVVAAVERTVRHG
TH_2659|M.thermoresistible__bu TTIFAIAAIERTIRK-
MSMEG_5311|M.smegmatis_MC2_155 GLVLLCSMGFRRGAEK
MAB_4214c|M.abscessus_ATCC_199 VITLVAALAVVQRRDV
: : .