For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MARIAEDLFLLLLDNASAQPGLDRRRRERVLGAAVLLDLAYMCRVRPALAGEPVPAGRLIALAGQFPPDP VADPAFEVLRRKPLTPQAALAKLGGPTQTHLESHLQAMGQIQRVRMPGKGFPGRGRYCWPLTNRDRVSGA RAALLAALFDGHNPTPPVAAIICLLHSVDGLGAVLSLNERGWRWVHARATEIATGSWVDENATALPEMNL AVTTSAVRPALIALLD
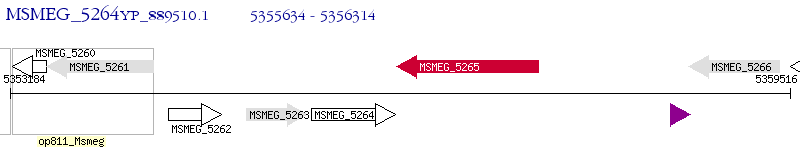
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5264 | - | - | 100% (226) | hypothetical protein MSMEG_5264 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2047 | - | 1e-67 | 60.71% (224) | hypothetical protein Mflv_2047 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1198c | - | 4e-42 | 42.79% (222) | hypothetical protein MAB_1198c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2170 | - | 2e-79 | 65.04% (226) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4669 | - | 5e-76 | 66.96% (224) | hypothetical protein Mvan_4669 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2047|M.gilvum_PYR-GCK MAQIAEDLFLLLLDNASAQPALDGARRGHVLAAASLLDLALDCRVRPAVD
Mvan_4669|M.vanbaalenii_PYR-1 MAQIAEDLFLLLVDNASAQPALDGSRREHVLAAAILLDLAHACRVRPAVT
MSMEG_5264|M.smegmatis_MC2_155 MARIAEDLFLLLLDNASAQPGLDRRRRERVLGAAVLLDLAYMCRVRPALA
TH_2170|M.thermoresistible__bu MAKIAEELFLLLLDNGSAQPALDRPRRTRVLTAAALLDLAYACRIRPAVP
MAB_1198c|M.abscessus_ATCC_199 MPQIAEDLLLLLLNNASGRPLLDKNRRSQALAAAIVLDLALAQRIRPATH
*.:***:*:***::*.*.:* ** ** :.* ** :**** *:***
Mflv_2047|M.gilvum_PYR-GCK GERAPAGRLVALDTEGP----VDPVAAPALAMLQSRPLRPGTAIKKLGRH
Mvan_4669|M.vanbaalenii_PYR-1 GEPVPEGRLVALDAPGP----LDPVSARAFALLRAKPLRPDAAIKKLGRD
MSMEG_5264|M.smegmatis_MC2_155 GEPVPAGRLIALAGQFP----PDPVADPAFEVLRRKPLTPQAALAKLGGP
TH_2170|M.thermoresistible__bu GEPVEAGRLLALSEPAPNAAPLDPVTEPVLRALRRQPLRPATVLTKLGKR
MAB_1198c|M.abscessus_ATCC_199 GEPTKAGHLLVLQAPDIG----DPVLDRAIHRLRRRPMDPTEAIAKVGRG
** . *:*:.* *** .: *: :*: * .: *:*
Mflv_2047|M.gilvum_PYR-GCK TEERLIAHLEQTGQIRRTVESTRRFG--KTVSYPLTGRGRVGPARQALTS
Mvan_4669|M.vanbaalenii_PYR-1 SQKLLVTSLTQAGQIRPTARPGKLFG--RSRSFPLTNRDRVGAARAALMA
MSMEG_5264|M.smegmatis_MC2_155 TQTHLESHLQAMGQIQRVRMPGKGFPGRGRYCWPLTNRDRVSGARAALLA
TH_2170|M.thermoresistible__bu TERNLRTHLEQTGEIRRIRVPGRRF--RREYAWPLTNRDRVGRARSDVLA
MAB_1198c|M.abscessus_ATCC_199 VESQMLHRLEITGDIHTVRMGGRLFP---DKYWPVTNNERANAVRQGVTD
: : * *:*: : * :*:*.. *.. .* :
Mflv_2047|M.gilvum_PYR-GCK ALFEGASPAPATAAVITVLHAVDGLGALLSLNDRGWRWVHARAGEIAGGS
Mvan_4669|M.vanbaalenii_PYR-1 ALFDRTPPAPATAAIITVLHAVDGLGALLSLNDRGWRWVHARAGEIASGS
MSMEG_5264|M.smegmatis_MC2_155 ALFDGHNPTPPVAAIICLLHSVDGLGAVLSLNERGWRWVHARATEIATGS
TH_2170|M.thermoresistible__bu ALFEGQVPAPSTAAIITLLHAVDGLGALLSLNDRGWRWVHARATEIATGA
MAB_1198c|M.abscessus_ATCC_199 VLFHYAPPSPSTAAIIAVLHGVNGFDAILSLDPIGKQQVSRWSQTIADGG
.**. *:*..**:* :**.*:*:.*:***: * : * : ** *.
Mflv_2047|M.gilvum_PYR-GCK WVDES-PSALPEVNLAVTAATVRQALAQLT-
Mvan_4669|M.vanbaalenii_PYR-1 WVDES-PTALPEVNLAVTASAVRQALVQLA-
MSMEG_5264|M.smegmatis_MC2_155 WVDEN-ATALPEMNLAVTTSAVRPALIALLD
TH_2170|M.thermoresistible__bu WVDEA-PSALPEMNLVVTTASVRPALM----
MAB_1198c|M.abscessus_ATCC_199 WAAQTRDNRGPAVNLAVTAAVISAALHSMS-
*. : . * :**.**:: : **